Abstract
Screening tests for disease have their performance measured through sensitivity and specificity, which inform how well the test can discriminate between those with and without the condition. Typically, high values for sensitivity and specificity are desired. These two measures of performance are unaffected by the outcome prevalence of the disease in the population. Research projects into the health of the American Indian frequently develop Machine learning algorithms as predictors of conditions in this population. In essence, these models serve as in silico screening tests for disease. A screening test’s sensitivity and specificity values, typically determined during the development of the test, inform on the performance at the population level and are not affected by the prevalence of disease. A screening test’s positive predictive value (PPV) is susceptible to the prevalence of the outcome. As the number of artificial intelligence and machine learning models flourish to predict disease outcomes, it is crucial to understand if the PPV values for these in silico methods suffer as traditional screening tests in a low prevalence outcome environment. The Strong Heart Study (SHS) is an epidemiological study of the American Indian and has been utilized in predictive models for health outcomes. We used data from the SHS focusing on the samples taken during Phases V and VI. Logistic Regression, Artificial Neural Network, and Random Forest were utilized as in silico screening tests within the SHS group. Their sensitivity, specificity, and PPV performance were assessed with health outcomes of varying prevalence within the SHS subjects. Although sensitivity and specificity remained high in these in silico screening tests, the PPVs’ values declined as the outcome’s prevalence became rare. Machine learning models used as in silico screening tests are subject to the same drawbacks as traditional screening tests when the outcome to be predicted is of low prevalence.
Impact statement
Artificial Intelligence (AI) and Machine Learning (ML) techniques are increasingly integrated into screening and diagnostic models to pinpoint individuals at risk of specific diseases or medical conditions. However, with the rise in popularity of AI and ML, the literature (and internet) is flooded with reports on computer-based prediction and screening tests, often focusing more on showcasing the technique rather than discussing their screening and diagnostic performance. In particular, there is a proliferation of algorithms created for minority groups, including the American Indian. A motivating factor in creating an in silico screening exam for American Indians is that this population, as a whole, experiences a greater burden of comorbidities, including diabetes mellitus, obesity, cancer, cardiovascular disease, and other chronic health conditions, than the rest of the U.S. population. This report evaluates these AI algorithms for the American Indian like a screening test in terms of performance in low prevalence situations.
Introduction
Artificial Intelligence (AI) and Machine Learning (ML) techniques are increasingly integrated into screening and diagnostic models to pinpoint individuals at risk for specific diseases or medical conditions [1]. However, with AI’s and ML’s rise in popularity, the literature (and the internet) is flooded with reports on computer-based prediction and screening tests, often focused more on showcasing techniques than discussing their screening and diagnostic performance. Advances in computer processing speed, increasing numbers of data scientists, low- to no-cost programming libraries, and availability of larger healthcare data sets have driven the proliferation of AI algorithms [2]. Kumar et al. have listed a sampling of prediction algorithms and data sets, including those for outcomes in Alzheimer’s disease, cancer, diabetes, chronic heart disease, tuberculosis, stroke, hypertension, skin disease, and liver disease, among others [3]. Notwithstanding the proliferation of algorithms, AI is positioned to considerably enhance the accuracy and efficiency of screening tests. Specifically, ML algorithms can be trained on extensive data sets to discern patterns and make predictive analyses based on those patterns. Rapid expansion of AI technology, coupled with enhanced computing power in health screening, underscores the necessity for evaluating the algorithm’s performance and quality of these algorithms [4].
The U.S. Government Accountability Office conducted a technology assessment noting that AI and ML offer advantages in analyzing underserved populations [5]. However, one challenge of utilizing AI in epidemiology pertains to the underrepresentation or absence of minority groups within these algorithms’ training data sets [6]. Also, screening test performance may vary in minority populations due to their differences in disease prevalence from non-minority populations.
Many AI and ML methods for predicting disease in non-minority populations are recalibrated for minority groups. For example, an ML algorithm for mortality prediction based on chronic disease was recalibrated for the population of South Korea; this adjusted index showed a greater mortality prediction than the original algorithm [7]. Another effort adjusted this mortality prediction algorithm using hospital discharge abstracts from six countries [8].
In terms of minority status, American Indians are sometimes referred to as the “minority of the minority” or the “invisible minority,” given their small population, cultural identity, languages, and histories that set them apart from other groups. Focusing on AI and ML can offer advantages in analyzing these underserved populations, who, like American Indians, bear a greater burden of certain health conditions [9–11]. This study focused on in silico AI and ML screening tests explicitly designed for the American Indian population.
The number of in silico diagnostic and screening tests has grown exponentially over the last decade, with many of these utilizing data sets based on American Indians. Our study serves as a reminder that in silico screening tests, even when classified as AI or ML algorithms, are still subject to the same limitations related to disease prevalence as those of their laboratory-based counterparts.
Popularization of Pima Indian data
Several research articles in the public domain report on AI and ML algorithms for diabetes classification in the Pima Indian population. A contributing factor is the availability of numerous Pima Indian data sets provided to the AI community through platforms like Kaggle, a popular resource for AI and ML algorithm developers [12]. However, these studies often overlooked the differences in disease prevalence among different populations and the potential consequences of applying algorithms trained specifically on one population to another.
Examples of ML algorithms for diabetes classification in Pima Indians sampled from the literature include Support Vector Machines, Radial Basis Function, Kernel Support Vector Machines, K-Nearest Neighbor, Artificial Neural Networks, Fuzzy Support Vector Machine, Naïve Bayes Classifier, J48 Decision Tree, and a Random Forest Classifier [13–15]. Some of the articles in this sample failed to recognize the high prevalence of diabetes among the Pima Indians and the impact of disease prevalence on screening test performance, and tended to focus solely on the methods used to perform the classifications [14].
The Strong Heart Study models
The Strong Heart Study (SHS) has played a significant role in identifying risk factors and patterns related to cardiovascular disease (CVD) in American Indian communities. It included 12 tribes located in Oklahoma, Arizona and the Dakotas. Statistical models developed using SHS data have informed interventions and public health policies targeting CVD. SHS data were also used in developing ML models and risk-based calculators addressing hypertension, diabetes, and coronary heart disease (CHD) [16–18].
AI and ML risks with American Indian data sets
However, potential risks are also associated with using ML in American Indian contexts. One notable concern involves the risk of ML algorithms perpetuating biases and stereotypes about American Indian communities. Specifically, algorithms trained on data sets that reinforce biases and stereotypes about American Indians could inadvertently foster further inequities against a population group frequently underrepresented in AI/ML training data sets for applications such as virtual screening tests. A lack of training data can also result in inaccuracies; a recent example involved an image recognition application identifying an American Indian in native dress as a bird [19]. To mitigate such risks, ML researchers and developers need to collaborate closely with American Indian communities to ensure their technologies are developed ethically and respectfully. Collaborations could entail establishing research partnerships with American Indian communities, involving community members in designing and developing ML models, and ensuring that models are built using unbiased and culturally sensitive data sets, like those of the SHS.
Traditional screening test performance
Traditional screening test performance is typically based on a gold standard in which an individual’s true disease status is known to establish the test’s sensitivity, specificity, positive predictive value (PPV), and negative predictive value (NPV). The test’s sensitivity and specificity inform its effectiveness in identifying the proportion of people in the population with and without the condition of interest [20]. Sensitivity is the ability of the test to correctly identify those with the condition, while specificity is the ability of the test to correctly identify those without it. Sensitivity can be calculated from the column of those truly positive for the condition, while specificity is derived from the column of those truly negative for the condition in Figure 1.
FIGURE 1
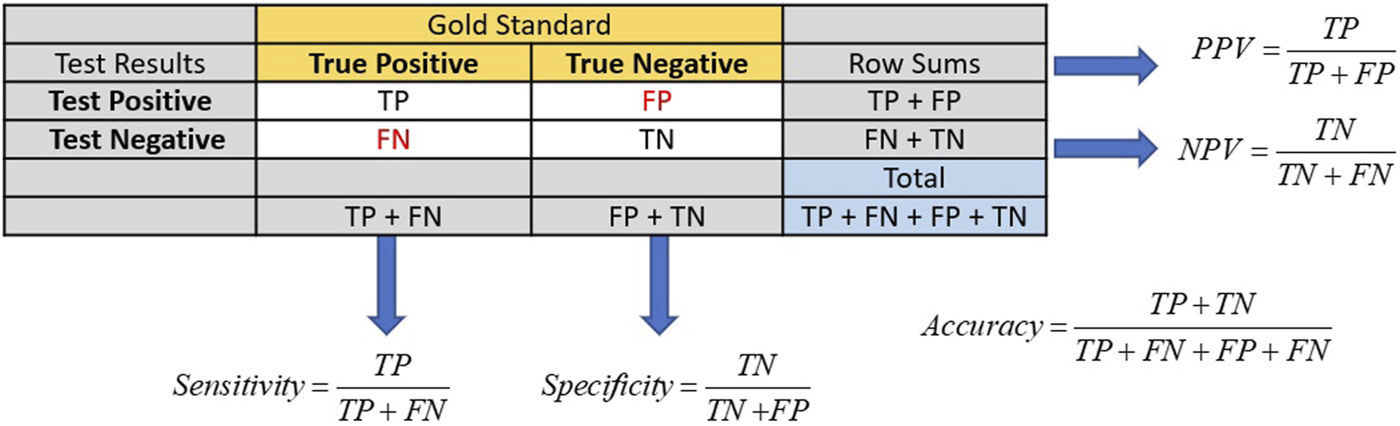
Calculations of sensitivity, specificity, PPV, and NPV for screening tests usually have their performance metrics determined via a gold standard. The numbers of true positives and negatives are represented by TP and TN, respectively. Likewise, the numbers of false positives and negatives are represented by FP and FN.
Among these metrics, the PPV holds clinical significance for both healthcare providers and patients. The PPV is a conditional probability that the tested individual has the disease, given that they tested positive. A high PPV indicates effective identification of individuals with the tested condition, guiding further testing, diagnosis, and treatment decisions. The PPV is calculated from the row in Figure 1 that represents those subjects who tested positive for disease.
As disease prevalence decreases, screening test performance decreases, particularly concerning the PPV. This decline can lead to situations where accuracy and sensitivity remain high, giving a false impression of a well-performing test due to the increased number of false positives (FP). For example, in a population of 1,000 people with a disease prevalence of 40%, a test with a sensitivity of 90% and specificity of 80% will produce a PPV of 75%. If the disease prevalence is lowered to 10% in this same population, the PPV drops to 33.33%; hence, the prevalence dominates in screening for rare diseases [21]. Therefore, healthcare providers should consider these factors when interpreting the PPV for further testing and treatment decisions.
While sensitivity and specificity provide information about test performance across populations, PPV is often more relevant in clinical practice. It helps physicians assess the likelihood of disease presence after a positive test result, especially in populations with low disease prevalence. If the disease outcome becomes increasingly rare, the algorithm will likely always predict the absence of disease, leading to high accuracy but poor PPV [22].
This study aimed to develop and evaluate three popular and commonly used AI and ML techniques as in silico screening tools for predicting three chronic conditions with differing prevalences in the SHS population: peripheral artery disease (PAD), hypertension, and type 2 diabetes. Specifically, we predicted the disease outcome using epidemiological data with methods including artificial neural networks (ANNs), random forest (RF), and logistic regression (LR). Unlike their traditional laboratory-based counterparts, these in silico tests do not have pre-determined sensitivity or specificity; rigorous testing has not been performed using a gold standard to establish these values. Our simulations provided a glimpse of the sensitivity, specificity, and PPV of these in silico screening tests, as these values changed in response to differing disease prevalences. We hypothesized that these in silico screening tools tailored to the American Indian population would show reduced performance as disease prevalence declines, regardless of the AI or ML method.
This research serves as a reminder that the limitations of screening tests regarding disease prevalence still apply, whether those tests are in silico AI or ML algorithms or traditional screening tools.
Materials and methods
LR, ANNs, and RFs are well-known methods for creating in silico screening tests. While RFs operate as a nonlinear model, LR requires a linear relationship with the regression coefficients. ANNs present a more intricate approach, often featuring multiple layers commonly known as deep learning. AI and ML can potentially enhance screening for various medical conditions by illuminating linear and nonlinear data relationships. Nonetheless, it is crucial to acknowledge that the application of AI in medical research and screening exams is still nascent, and concerns over AI algorithms’ accuracy and reliability linger.
Longitudinal epidemiological SHS data set
The SHS began in 1988 as a multi-center, population-based longitudinal study of cardiovascular disease (CVD) and its risk factors among American Indians. The study had three phases: a clinical examination, a personal interview, and an ongoing mortality and morbidity survey [23]. Participants from 12 different American Indian tribes were recruited from Arizona, Oklahoma, and the Dakotas, aided by volunteers from each community who promoted participation [24].
Phase II of the SHS, examining changes in risk factors for CVD in the original cohort, occurred between 1993 and 1995. The Strong Heart Family Study (SHFS), launched in Phase III (1998–1999), investigated genetic determinants of cardiovascular disease and extended recruitment to the original cohort’s family members aged 18 years and older [25]. Phase IV (2001–2003) involved surveillance of the original cohort plus 90 families to continue the study of genetic markers for CVD [26]. The Phase V exam (2006–2009) continued the SHFS, which began in Phase III; all participants from Phase III and IV were invited to participate in examinations conducted at local Indian Health Service hospitals, clinics, or tribal community facilities [27]. In Phase VI (2014–2018), all surviving participants were invited to complete a medical questionnaire, and continued the morbidity and mortality surveillance continued.
Physicians on the SHS Morbidity and Mortality (M & M) review committee examined the types of health-related events requiring hospital treatment and subsequent causes of mortality, when it occurred. Two of these physicians independently reviewed fatal events for cause, with the results reconciled by a third physician. In addition, one physician reviewed the medical records regarding study participant’s non-fatal events to verify specific diagnoses (i.e., stroke). This surveillance occurred yearly for both the original cohort and family cohort participants.
The available 2,468 Phase V SHS participants were divided into development and training cohorts (80%), while the remaining sample (20%) was assigned to a testing cohort. The training cohort generated the model weights, while the testing cohort assessed the algorithm’s quality.
A 1-year time-to-event data set for this study was constructed from the examination date in Phase V. The M & M results and all Phases of SHS data will cumulatively provide information on the subject’s medical conditions and mortality outcomes. Basic descriptive demographic statistics by gender, age, and comorbidity, including the numbers and percentages for binary variables, are listed in Table 1.
TABLE 1
| Medical condition | All | Male | Female |
|---|---|---|---|
| N (%) | 2,468 | 977 (39.59) | 1,491 (60.41) |
| Age (years) | |||
| Mean (SD) | 45.55 (16.41) | 43.74 (16.00) | 46.73 (16.58) |
| Median | 44.40 | 42.70 | 45.70 |
| Hypertension (%) | 948 (38.41) | 402 (42.41) | 546 (57.59) |
| Diabetes (%) | 631 (25.56) | 240 (38.03) | 391 (61.97) |
| PAD (%) | 94 (3.81) | 32 (34.04) | 62 (65.96) |
Age, gender, and medical condition of SHS Phase V participants.
Data labels for hypertension and diabetes already existed within the SHS Phase V data set but did not include a specific label for PAD. The data included the participants’ right and left ankle-brachial indexes (ABIs), which were used to define the presence or absence of PAD. This study used a resting ABI of less than 0.90 on either the right, left, or both sides, similar to that in Virane et al., to indicate a PAD diagnosis. Participants were coded as either 1 or 0 for the presence or absence of PAD, respectively [28, 29].
LR, ANN, and RF were then used to model the PAD, hypertension, and diabetes target features. These models ran 100 unique iterations of splitting and training the data, and producing metrics from the test set. Metrics tracked for the models were accuracy, specificity, sensitivity, PPV, and NPV, which were averaged over 100 iterations for each model type.
SAS version 9.4 was used to assemble the Phases of the SHS into a single data set, while Python version 3.9.7 was used to script the LR, RF, and ANN models.
Results
Numbers of SHS participants reporting hypertension, diabetes, and positivity for PAD are reported in Table 1.
More females than males participated in Phase V, comprising over 60% of the study participants. In addition, women reported higher percentages of hypertension, diabetes, and PAD than did men.
Figures 2–4 show each model’s accuracy, sensitivity, specificity, NPV, and PPV and reflect similar performance patterns among all models. The PPV and sensitivity measures seem to suffer the most as the outcome prevalence declines, which is what is typically observed for a traditional laboratory-based screening test. PPV and sensitivity decline for all models but remain parallel for LR and appear to converge within the ANN and RF models. As PPV and sensitivity decline for the RF model, they converge to zero at an outcome prevalence of 4% (PAD). Specific numerical values for each model metric are recorded in Table 2 for all three chronic conditions.
FIGURE 2
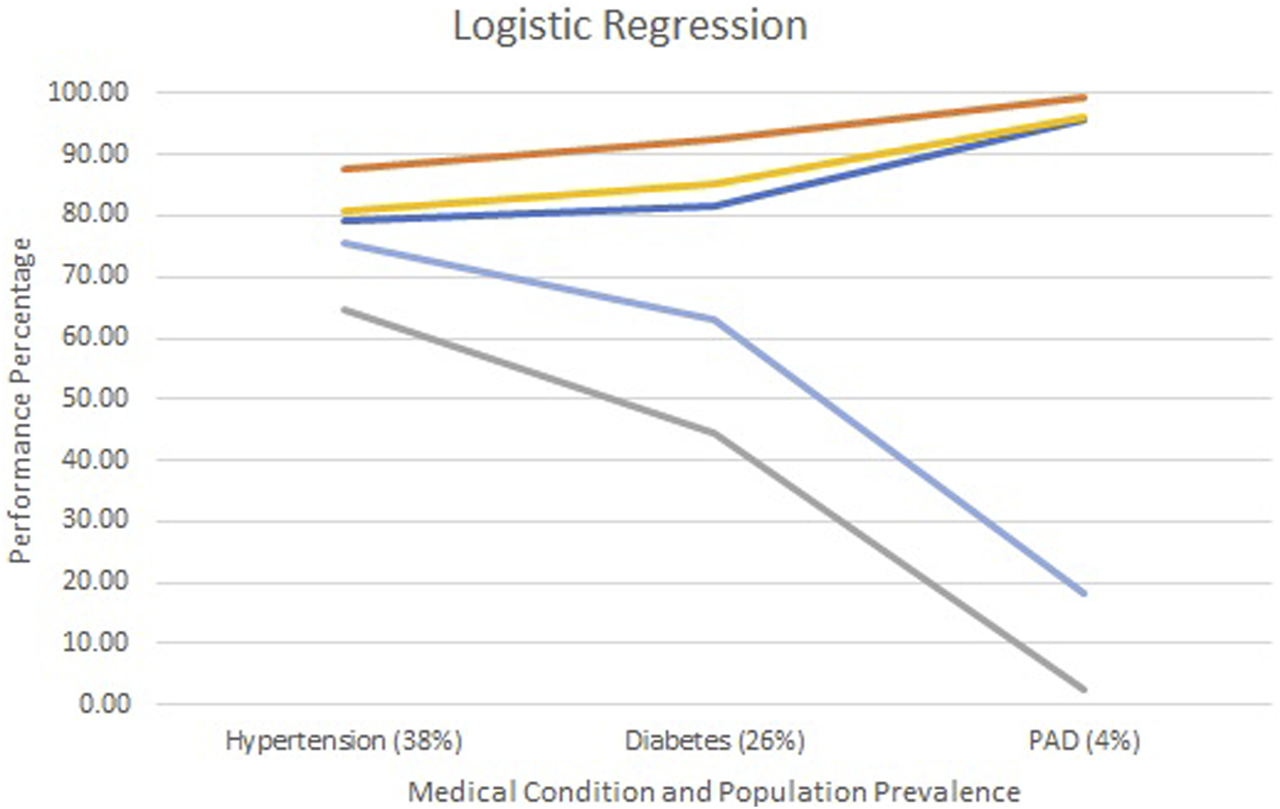
Screening test diagnostics for logistic regression.  Accuracy
Accuracy  Specificity
Specificity  Sensitivity
Sensitivity  PPV
PPV  NPV.
NPV.
FIGURE 3
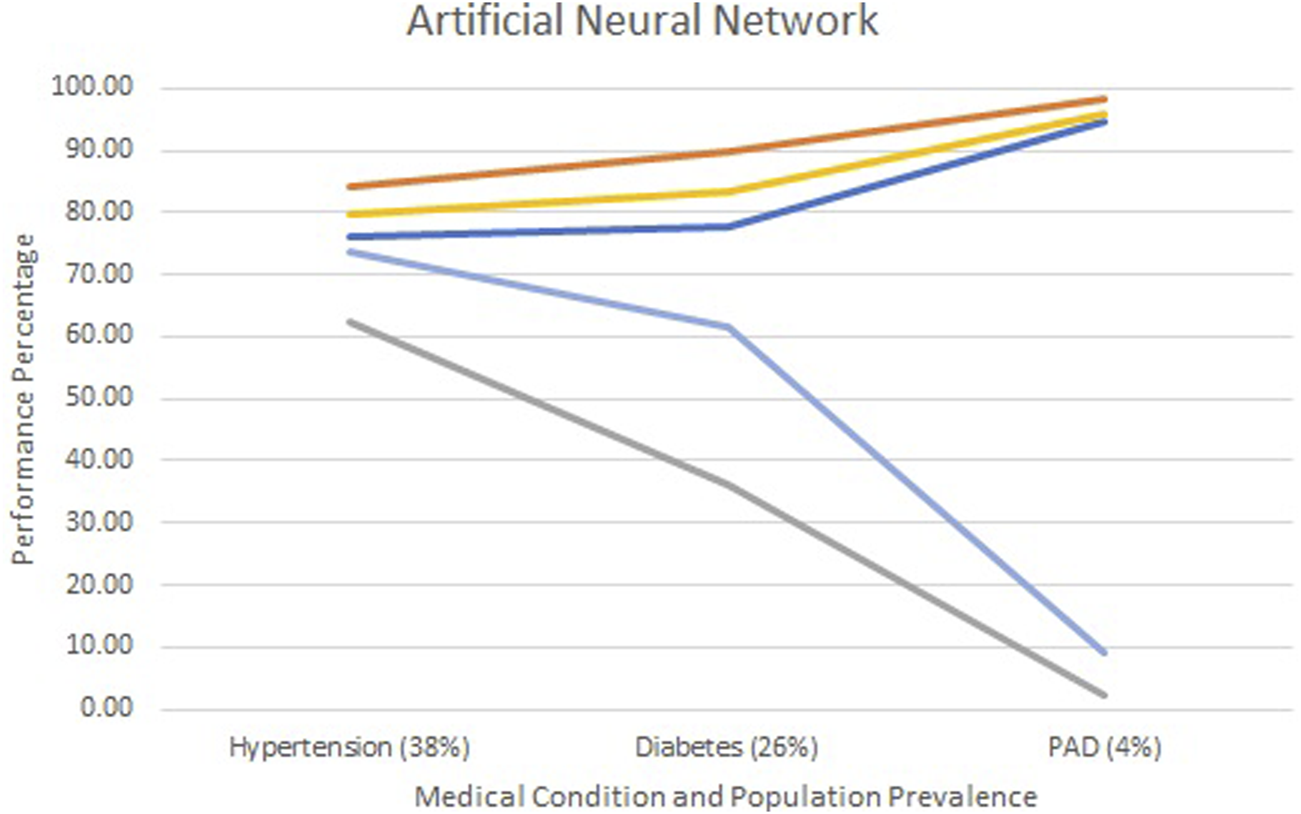
Screening test diagnostics for artificial neural networks.  Accuracy
Accuracy  Specificity
Specificity  Sensitivity
Sensitivity  PPV
PPV  NPV.
NPV.
FIGURE 4
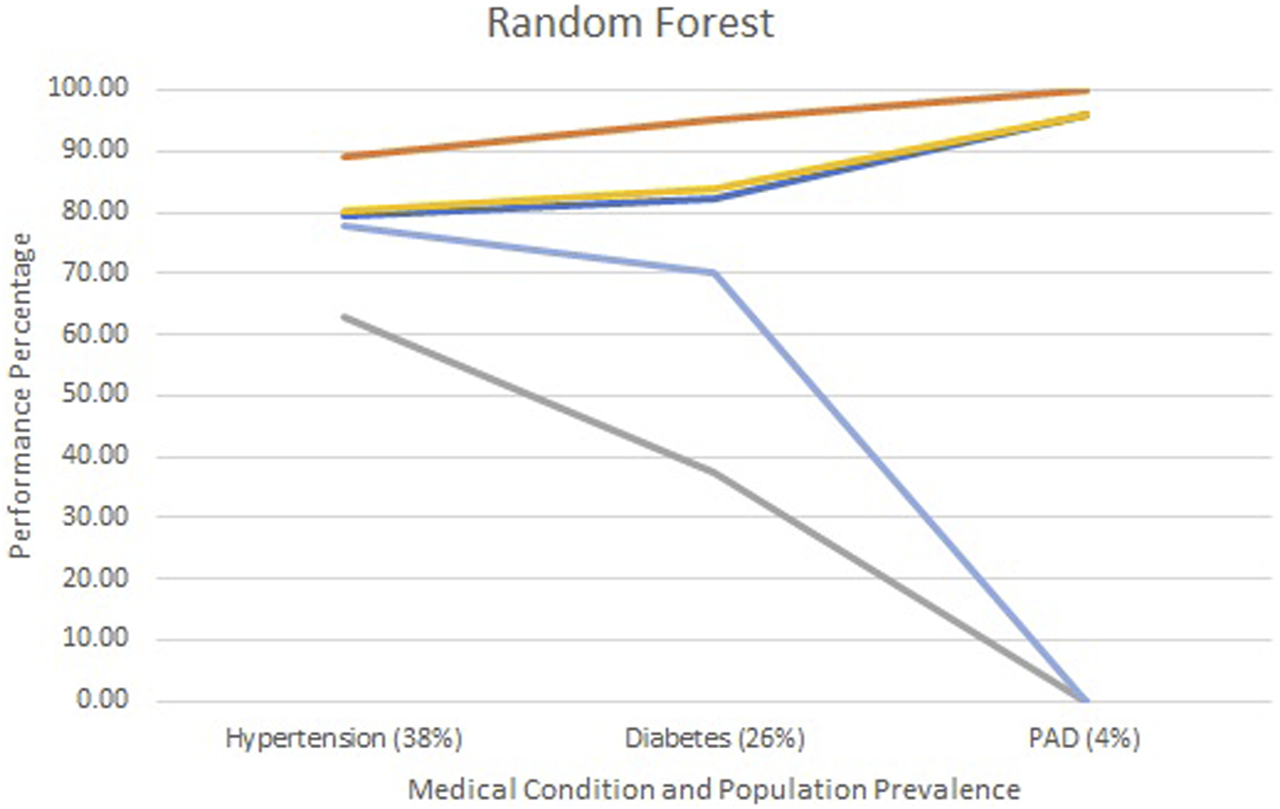
Screening test diagnostics for random forest.  Accuracy
Accuracy  Specificity
Specificity  Sensitivity
Sensitivity  PPV
PPV  NPV.
NPV.
TABLE 2
| Metric | Model and chronic condition | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Hypertension | Diabetes | PAD | |||||||
| LR | ANN | RF | LR | ANN | RF | LR | ANN | RF | |
| Accuracy | 78.99 | 76.00 | 79.35 | 81.49 | 77.66 | 82.18 | 95.57 | 94.56 | 95.99 |
| Specificity | 87.52 | 84.21 | 89.24 | 92.28 | 89.92 | 95.31 | 99.47 | 98.44 | 100.00 |
| Sensitivity | 64.79 | 62.41 | 62.88 | 44.59 | 36.06 | 37.26 | 2.40 | 2.07 | 0.00 |
| PPV | 75.61 | 73.62 | 77.75 | 62.88 | 61.77 | 70.13 | 18.06 | 9.04 | 0.00 |
| NPV | 80.63 | 79.87 | 80.12 | 85.08 | 83.52 | 83.88 | 96.06 | 96.00 | 95.99 |
Summary of screening test diagnostics by method: logistic regression, artificial neural network, and random forest are represented by LR, ANN, and RF.
All three models reported accuracy and specificity values that increased as the condition’s prevalence declined. These two measures are roughly 95% or higher for PAD, regardless of the model selected. Conversely, sensitivity and PPV decreased as the prevalence declined, largely due to the increased number of false positives. Although poor, the LR model reported the greatest PPV of 18% for PAD, as compared to the ANN and RF, which were at 9% and 0%, respectively.
The formulas for sensitivity and PPV in Figure 1 give insight to the effect of false and true positives on these two metrics. Traditional laboratory screening tests’ performance metrics are usually determined via a gold standard. As the prevalence of the condition declined, so did the number of true positives, while that of false positives increased, driving down both the sensitivity and PPV. Accuracy remained high as the true negatives grew, inflating these metrics.
Discussion
LR, ANNs, and RFs are popular methods in the burgeoning world of AI and ML. Although these methods are quite different from one another, we can see that their performance metric trends are similar in screening for disease outcomes with varying prevalences. These performance metrics give the developers of these methods an idea of how a specific in silico screening method will perform in the population it was designed to serve based on the prevalence of the outcome.
Although these algorithms may have high predictive power, as measured in terms of predictive accuracy, some are criticized for lacking any causal reasoning [30]. For example, ANNs may give reliable predictions for the end users; however, these end users do not know how the algorithm came to a particular conclusion. Thus, they are “black boxes” contributing little to understanding a condition’s cause.
Regardless of the method used, the PPV declined in parallel with the overall prevalence of the condition. The type of in silico modeling approach is still subject to the same limitations as those of traditional lab-based screening tests, an important factor to remember as online screening tests become more widespread. This study reminds us that regardless of the approach used, in silico AI and ML screening tests are not “magic bullets.” Their performance is still limited by the prevalence of the disease in the populations they are intended to serve.
Statements
Data availability statement
Publicly available datasets were analyzed in this study. This data can be found here: https://strongheartstudy.org/.
Ethics statement
This project was approved by the University of Oklahoma Health Sciences Center Institutional Review Board along with the Strong Heart Study Publications and Presentations Committee (SHS700). In addition, the National Center for Toxicological Research Institutional Review Board approved the project and its publication.
Author contributions
TM performed the modeling and Python coding, while PR completed the statistical analysis and initial manuscript writing. DW and WT contributed to the structure and content of the manuscript. YZ and JR described the Strong Heart Study design and contributed to the creation of the manuscript. All authors contributed to the article and approved the submitted version.
Funding
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. The Strong Heart Study has been funded in whole or in part with federal funds from the National Heart, Lung, and Blood Institute, National Institutes of Health, Department of Health and Human Services, under contract numbers (75N92019D00027, 75N92019D00028, 75N92019D00029, and 75N92019D00030). The study was previously supported by research grants: R01HL109315, R01HL109301, R01HL109284, R01HL109282, and R01HL109319 and by cooperative agreements: U01HL41642, U01HL41652, U01HL41654, U01HL65520, and U01HL65521.
Conflict of interest
The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Author disclaimer
This manuscript reflects the views of its authors and does not necessarily reflect those of the U.S. Food and Drug Administration. Any mention of commercial products is for clarification only and is not intended as approval, endorsement, or recommendation.
References
1.
Davenport T Kalakota R . The potential for artificial intelligence in healthcare. Future Healthc J (2019) 6:94–8. 10.7861/futurehosp.6-2-94
2.
Bohr A Memarzadeh K . The rise of artificial intelligence in healthcare applications. In: BohrAMemarzadehK, editors Artificial intelligence in healthcare. Academic Press (2020). p. i–iii.
3.
Kumar Y Koul A Singla R Ijaz MF . Artificial intelligence in disease diagnosis: a systematic literature review, synthesizing framework and future research agenda. J Ambient Intelligence Humanized Comput (2022) 14:8459–86. 10.1007/s12652-021-03612-z
4.
Chalkidou A Shokraneh F Kijauskaite G Taylor-Phillips S Halligan S Wilkinson L et al Recommendations for the development and use of imaging test sets to investigate the test performance of artificial intelligence in health screening. Lancet Digital Health (2022) 4:e899–e905. 10.1016/S2589-7500(22)00186-8
5.
U. S. Government Accountability Office. Artificial intelligence in health care: benefits and challenges of machine learning technologies for medical diagnostics (2024). Available from: https://www.gao.gov/products/gao-22-104629 (Accessed July 1, 2024).
6.
Sung J Hopper JL . Co-evolution of epidemiology and artificial intelligence: challenges and opportunities. Int J Epidemiol (2023) 52:969–73. 10.1093/ije/dyad089
7.
Choi JS Kim MH Kim YC Lim YH Bae HJ Kim DK et al Recalibration and validation of the Charlson comorbidity index in an Asian population: the national health insurance service-national sample cohort study. Sci Rep (2020) 10:13715. 10.1038/s41598-020-70624-8
8.
Quan H Li B Couris CM Fushimi K Graham P Hider P et al Updating and validating the Charlson comorbidity index and score for risk adjustment in hospital discharge abstracts using data from 6 countries. Am J Epidemiol (2011) 173:676–82. 10.1093/aje/kwq433
9.
Benjamin EJ Muntner P Alonso A Bittencourt MS Callaway CW Carson AP et al Heart disease and stroke statistics-2019 update: a report from the American heart association. Circulation (2019) 139:e56–e528. 10.1161/CIR.0000000000000659
10.
Centers for Disease Control and Prevention. CDC and Indian country working together (2024). Available from: https://stacks.cdc.gov/view/cdc/44668 (Accessed July 1, 2024).
11.
Indian health Service. Disparities (2024). Available from: https://www.ihs.gov/newsroom/factsheets/disparities/ (Accessed July 1, 2024).
12.
Kaggle. Kaggle: your home for data science (2023). Available from: https://www.kaggle.com/ (Accessed April 28, 2023).
13.
Chang V Bailey J Xu QA Sun Z . Pima Indians diabetes mellitus classification based on machine learning (ML) algorithms. Neural Comput Appl (2022) 35:16157–73. 10.1007/s00521-022-07049-z
14.
Kaur H Kumari V . Predictive modelling and analytics for diabetes using a machine learning approach. Appl Comput Inform (2022) 18:90–100. 10.1016/jaci2018.12.004
15.
Lukmanto R Irwansyah E . The early detection of diabetes mellitus (DM) using Fuzzy hierarchical model. Proced Comput Sci (2015) 59:312–9. 10.1016/j.procs.2015.07.571
16.
Lee ET Howard BV Wang W Welty TK Galloway JM Best LG et al Prediction of coronary heart disease in a population with high prevalence of diabetes and albuminuria: the Strong Heart Study. Circulation (2006) 113:2897–905. 10.1161/CIRCULATIONAHA.105.593178
17.
Wang W Lee ET Fabsitz RR Devereux R Best L Welty TK et al A longitudinal study of hypertension risk factors and their relation to cardiovascular disease: the Strong Heart Study. Hypertension (2006) 47:403–9. 10.1161/01.HYP.0000200710.29498.80
18.
Wang W Lee ET Howard BV Fabsitz RR Devereux RB Welty TK . Fasting plasma glucose and hemoglobin A1c in identifying and predicting diabetes: the strong heart study. Diabetes Care (2011) 34:363–8. 10.2337/dc10-1680
19.
Cipolle AV . How native Americans are trying to debug A.I.’s Biases (2024). Available from: https://www.nytimes.com/2022/03/22/technology/ai-data-indigenous-ivow.html (Accessed July 1, 2024).
20.
Gordis L . Epidemiology. Philadelphia: Elsevier Saunders (2004). p. 335.
21.
van Belle G . Statistical rules of thumb. 2nd ed.Hoboken: Basic Books, Inc (2008). p. 272.
22.
Yu Z Wang K Wan Z Xie S Lv Z . Popular deep learning algorithms for disease prediction: a review. Cluster Comput (2023) 26:1231–51. 10.1007/s10586-022-03707-y
23.
Lee ET Welty TK Fabsitz R Cowan LD Le NA Oopik AJ et al The Strong Heart Study. A study of cardiovascular disease in American Indians: design and methods. Am J Epidemiol (1990) 132:1141–55. 10.1093/oxfordjournals.aje.a115757
24.
Stoddart ML Jarvis B Blake B Fabsitz RR Howard BV Lee ET et al Recruitment of American Indians in epidemiologic research: the strong heart study. Am Indian Alsk Native Ment Health Res (2000) 9:20–37. 10.5820/aian.0903.2000.20
25.
North KE Howard BV Welty TK Best LG Lee ET Yeh JL et al Genetic and environmental contributions to cardiovascular disease risk in American Indians: the strong heart family study. Am J Epidemiol (2003) 157:303–14. 10.1093/aje/kwf208
26.
Strong Heart Study. Strong heart study phase IV operations manual (2024). Available from: https://strongheartstudy.org/portals/1288/Assets/documents/manuals/Phase%20IV%20Operations%20Manual.pdf?ver=2017-11-15-134610-080 (Accessed July 1, 2024).
27.
Strong Heart Study. Strong heart study phase IV operations manual (2024). Available from: https://strongheartstudy.org/portals/1288/Assets/documents/manuals/Phase%20V%20Operations%20Manual.pdf?ver=2017-11-15-134617-657 (Accessed July 1, 2024).
28.
American Heart Association. How is PAD diagnosed (2024). Available from: https://www.heart.org/en/health-topics/peripheral-artery-disease/diagnosing-pad (Accessed July 1, 2024).
29.
Virani SS Alonso A Benjamin EJ Bittencourt MS Callaway CW Carson AP et al Heart disease and stroke statistics-2020 update: a report from the American heart association. Circulation (2020) 141:e139–e596. 10.1161/CIR.0000000000000757
30.
Pearl J Mackenzie D . The book of why: the new science of cause and effect. Basic Books, Inc. (2018). p. 432.
Summary
Keywords
artificial intelligence, machine learning, screening test, American Indian, low prevalence
Citation
Rogers P, McCall T, Zhang Y, Reese J, Wang D and Tong W (2025) Leveraging AI to improve disease screening among American Indians: insights from the Strong Heart Study. Exp. Biol. Med. 249:10341. doi: 10.3389/ebm.2024.10341
Received
09 August 2024
Accepted
16 December 2024
Published
08 January 2025
Volume
249 - 2024
Updates
Copyright
© 2025 Rogers, McCall, Zhang, Reese, Wang and Tong.
This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Paul Rogers, paul.rogers@fda.hhs.gov
Disclaimer
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.