- 1National Medical Research Center of Cardiology Named After E. I. Chazov, Moscow, Russia
- 2Medical Research and Education Center Lomonosov Moscow State University, Moscow, Russia
Abstract
Hypertrophic cardiomyopathy (HCM) is a genetic cardiac disorder associated with an increased risk of arrhythmias, heart failure, and sudden cardiac death. Current imaging and clinical markers are not fully sufficient in accurate diagnosis and patient risk stratification. Although known cardiac biomarkers in blood are used, they lack specificity for HCM and primarily stratify for death due to heart failure in overt cases. Non-coding RNAs, particularly microRNAs, have emerged as promising biomarkers due to their role in regulating gene expression in both healthy and pathological hearts. Circulating microRNA signatures may dynamically reflect the progression of HCM, offering potential utility in diagnosis and disease monitoring as well as inform biologic pathways for innovative therapeutic strategies. However, studying microRNAs in cardiovascular diseases is still in its early stages and poses many challenges. This review focuses on emerging research perspectives using advanced cardiac magnetic resonance techniques. We presume, that the search for circulating miR signatures associated with specific adverse myocardial features observed on cardiac magnetic resonance imaging - such as fibrosis, disarray, and microvascular disease - represents a promising direction in HCM research.
Impact statement
We are pleased to submit our manuscript, which highlights a perspective direction in hypertrophic cardiomyopathy research. Our study focuses on non-coding RNAs, specifically microRNAs, as promising cardiac biomarkers, and advanced cardiac magnetic resonance (CMR) imaging as a research tool that could facilitate the discovery of novel circulating miR biomarkers. The current biomarkers are not specific for HCM, and their use is limited by risk stratification for heart failure death. By reviewing recent literature, we discuss the potential to identify specific circulating microRNA signatures linked to adverse microanatomical features of HCM observed using advanced CMR. We aim to engage the HCM scientific community in future interdisciplinary collaborations. The brief review of evolving modalities already applied in some areas of clinical practice may be of interest to a broad audience of practitioners, including cardiologists, radiologists, laboratory specialists, and genetics.
Introduction
Hypertrophic cardiomyopathy (HCM) is the most common genetic cardiac disorder, characterized by left ventricular (LV) hypertrophy in the absence of abnormal loading conditions [1]. In addition to cardiomyocyte hypertrophy, histological hallmarks include myocardial fibrosis (focal and diffuse), extensive disarray, and microvascular disease (MVD), all linked to arrhythmias, sudden cardiac death (SCD), and heart failure [2–6]. HCM exhibits significant heterogeneity in LV morphology [7], clinical course [8], and genetic etiology, involving both monogenic and polygenic components. Over recent decades, genetic studies have focused on protein-coding genes, identifying sarcomeric gene variants as the primary cause in approximately half of HCM patients [9–11]. This has established sarcomere dysfunction as a crucial mechanism in HCM [12], prompting the development of new treatments, such as cardiac myosin inhibitors [13]. Despite notable advancements, challenges remain in altering disease progression [14]. In clinical practice, there is a need for disease-specific plasma biomarkers to differentiate HCM from secondary LV hypertrophy, enhance risk stratification, and monitor phenotype evolution in preclinical mutation carriers. A fundamental question is how to identify the signaling pathways and regulatory networks that mediate the phenotypic expression of HCM’s complex genetics.
This paper reviews micro non-coding ribonucleic acids (microRNAs or miRs), known as negative controllers of gene expression, as promising biomarkers for HCM. A special focus is placed on a) myocardial microanatomical features of HCM as a research field for disease-specific biomarkers; b) whether cardiac magnetic resonance (CMR) tissue characterization techniques hold potential to advance the discovery of circulating biomarkers, such as miRs, in HCM (Figure 1).
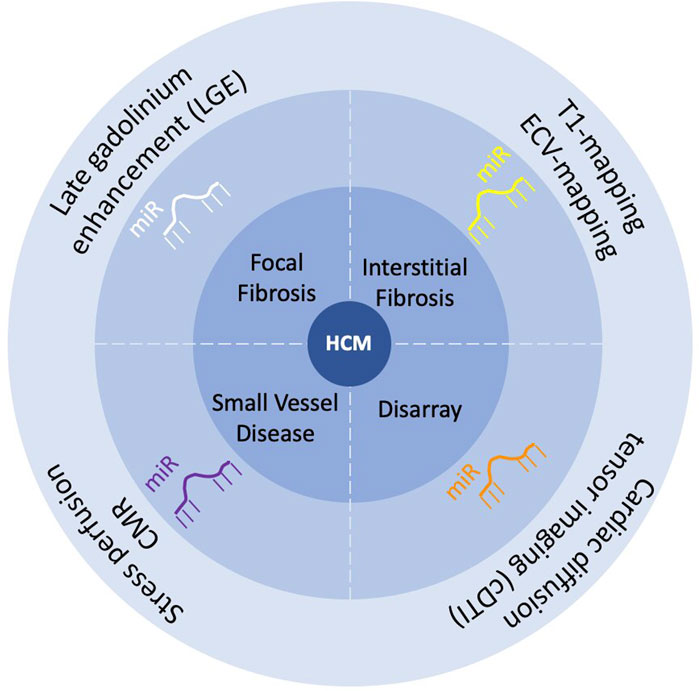
Figure 1. Scheme of the review “Circulating microRNA as promising biomarkers in hypertrophic cardiomyopathy: Can advanced cardiac magnetic resonance unlock new insights in research?”.
Current biomarkers for HCM
Since myocardial biopsy is not typically used in managing HCM, biomarkers are sought from cardiac imaging, clinical features, and circulating molecules. Similar to other cardiovascular diseases, a multi-parametric approach is employed for the diagnosis and risk stratification in HCM.
The only validated diagnostic marker for HCM is LV myocardial thickness, defined as ≥15 mm in adult probands and routinely assessed using echocardiography or CMR [1]. However, this marker has limitations: it cannot always reliably distinguish HCM from secondary LV hypertrophy or metabolic disorders and is insufficient to detect preclinical and early-stage disease. Furthermore, the quality of echocardiographic image may be compromised by patient-related factors, while CMR, although more precise, is not universally accessible and has procedural restrictions. Supporting diagnostic tools include electrocardiogram, family screening, genetic testing, and clinical assessment to exclude phenocopies. Currently, there are no specific blood biomarkers for HCM. While several molecules reflecting myocardial wall stretch, fibrosis, inflammation, apoptosis, necrosis, and endothelial dysfunction have demonstrated correlations with imaging findings [15], they have not been integrated into clinical practice due to insufficient evidence of specificity, thereby limiting their diagnostic utility. Moreover, none of these biomarkers exhibit adequate sensitivity to detect subclinical HCM [16].
Few imaging markers and clinical features - such as ventricular tachycardia, unexplained syncope, and a family history of SCD - are incorporated into SCD risk stratification algorithms [17, 18]. However, these models do not include blood biomarkers and show modest predictive value. Current European guidelines recommend the use of N-terminal pro-B-type natriuretic peptide (NT-proBNP) and troponin T for assessing mortality risk in overt HCM, particularly for death due to heart failure [1]. Additionally, NT-proBNP can serve as a surrogate marker for evaluating the therapeutic efficacy of cardiac myosin inhibitors [19, 20]. However, no risk models are currently designed to predict heart failure or other non-SCD-related mortality in HCM.
Challenges in identifying blood biomarkers for HCM may be attributed to historically small sample sizes in cardiomyopathy studies, inaccurate phenotype characterization, and truly weak biomarker correlations. Furthermore, simplistic approach based on a limited set of pre-selected biomarkers may not capture the complex underlying pathways.
Recent advancements in omics technologies, enabling the quantification of thousands of low-abundance circulating molecules (e.g., RNA, proteins, metabolites), have led to the discovery of novel biomarkers. Studies have demonstrated the ability of compiled protein and miR panels to differentiate HCM from hypertensive heart disease [21, 22] and subclinical HCM from controls [23, 24].
Circulating ncRNAs as biomarkers: case of miRs
Advances in cardiovascular genetics have shifted scientific interest toward the non-protein-coding genome and its non-coding RNA (ncRNA) transcripts [25]. The foremost function of ncRNAs is to regulate protein-coding gene expression, thereby affecting fundamental processes such as growth, differentiation, metabolism, apoptosis, and autophagy [26–29]. With growing evidence on the involvement of ncRNAs in heart diseases [30] and the advent of advanced computational methods, researchers can now explore the diagnostic, prognostic, and therapeutic potential of ncRNAs as biomarkers and novel targets for intervention [31].
Among various types of ncRNAs [32], miRs are the most abundant [33] and of particular interest in human diseases, including HCM [34]. MiRs are small molecules (∼20–22 nucleotides) that form a coordinated regulatory system and control gene expression post-transcriptionally by binding to their messenger RNAs, resulting in cleavage or translational repression [35]. A single miR can have numerous high- and low-affinity gene targets, and a single gene can be regulated by multiple miRs.
MiRs are detected not only in tissues but also in the bloodstream, making them attractive biomarkers for cardiac diseases, where biopsy is uncommon. They enter circulation from living and dead cells via active secretion or passive release. Despite the RNase-rich environment of blood, circulating miRs remain stable [36], protected within extracellular vesicles or bound to RNA-binding proteins [37, 38]. As conserved regulators of gene expression, miRs serve as dynamic biomarkers that reflect disease stages [39]. In heart failure studies, circulating miRs significantly improved the diagnostic power of NT-proBNP, which may be particularly beneficial for identifying heart failure with preserved ejection fraction, where standard clinical assessment, imaging, and NT-proBNP alone may not be definitive [40, 41].
MiRs in HCM
Studies in animal and human tissues indicate that miRs significantly influence HCM [39, 42–48]. Several miRs, such as miR-1, -21, -30b, -132, -133a, -133b, -150, -199a-3p, and -486-3p, consistently show altered expression across at least two independent studies, suggesting a role in HCM development [39, 45, 47]. Plasma miR levels may reflect myocardial pathology, and around 30 circulating miRs have been identified as potential HCM biomarkers. However, the diagnostic accuracy of individual circulating miRs remains moderate [34].
Recent research has expanded miR panels to improve diagnostic accuracy. A six-miR set, including miR-181a-5p, -181c-5p, -328-3p, -301a-3p, -193b-3p, and -142-3p, outperformed individual miRs and differentiated sarcomeric variant carriers with and without the HCM phenotype with high statistical significance [23]. In a larger study involving 555 patients, transcriptomic profiling of 1,141 miRs identified a panel of 20+ circulating miRs that effectively discriminated HCM from hypertensive LV hypertrophy. Subsequent pathway analysis linked these miRs to key signaling pathways, including Ras-MAPK [22].
In our study, patients harbouring disease-causing MYH7 variants had significantly higher plasma levels of miR-499a-5p compared to those with other sarcomeric variants, genotype-negative patients, and healthy controls [49]. MiR-499 is part of the “myo-miRs,” encoded by introns of cardiac myosin genes, including MYH7. These genes regulate muscle function by controlling the expression of both contractile proteins and regulatory miRs [50]. This finding supports a gene-oriented approach to studying miRs, as different genetic backgrounds may lead to distinct miR profiles that influence the disease phenotype.
Limitations of circulating miRs for biomarkers
While circulating miRs hold promise as biomarkers, several challenges must be addressed before their clinical application. Unlike miRs measured in tissues, which can be cell-type-specific [51], circulating miRs often do not provide clear tissue- or disease-specific signals. Of the 2,600+ miRs identified, only a few - such as miR-1, -133a, -208a/b, and -499 - are categorized as cardiac-specific [52]. Inconsistencies in miR profiles between studies complicate their reliability as biomarkers. This variability may stem from differences in cohort characteristics, methodological processes, or the complex nature of miR regulation, where miRs typically have modest effects on many targets rather than having a dramatic impact on single genes. However, the combined effects of miRs on targets within a shared pathway can be synergistic [53], suggesting that miR panels, rather than individual miRs, could enhance diagnostic accuracy. Further challenges and solutions are detailed elsewhere [34, 54].
Microanatomical features of HCM as a research field
Microanatomical changes in the myocardium, such as fibrosis, disarray, and MVD, are closely associated with HCM and its major clinical outcomes, as demonstrated in early histological studies [2–6], making them attractive substrates for the discovery of non-invasive biomarkers. Importantly, these changes are not merely secondary to LV hypertrophy but are also present in non-hypertrophied LV segments [55] and at the preclinical stage [56–58]. In mouse models, early administration of mavacamten, a first-in-class cardiac myosin inhibitor, suppressed the development of myocardial disarray and fibrosis by attenuating hypertrophic and profibrotic gene expression [59]. This highlights the role of specific signaling pathways in these adverse microanatomical changes in HCM. The identification of miR signatures associated with these features appears to be a promising avenue for HCM research.
Non-invasive myocardial characterization with CMR imaging
CMR tissue characterization techniques offer a non-invasive, radiation-free assessment of fibrosis, microstructure, and microvascular health in HCM, with ongoing research improving their clinical utility.
Late gadolinium enhancement (LGE) is widely used to demonstrate replacement fibrosis, indicating scar tissue resulting from cardiomyocyte death. In contrast, interstitial fibrosis, which represents increased extracellular matrix and volume (ECV) without the pre-requisite cardiomyocyte death, is best evaluated using native T1-mapping and ECV measurements [60]. Both LGE (2-standard deviation technique) and T1/ECV-mapping have been histologically validated to accurately reflect myocardial fibrosis and collagen volume [6, 61–65]. Cardiac diffusion tensor imaging (cDTI) is an innovative CMR technique that assesses myocardial microstructure by mapping water diffusion along muscle fibers [66]. This method may reveal myocyte disarray, as demonstrated in preclinical models [67], although histological validation in humans is still needed. MVD can be assessed through perfusion CMR imaging, which quantifies myocardial blood flow, myocardial perfusion reserve, and perfusion defects [68]. It is important to adjust perfusion maps for LGE, as LGE contributes to resting perfusion defects in 30% of patients, potentially confounding the assessment of ischemic burden in HCM [69].
Although CMR-derived tissue parameters display specific features, they are not pathognomonic for HCM and can also be seen in other conditions. Caution is needed, as, for instance, markers like T1 relaxation time and ECV may reflect amyloidosis or edema. Tissue findings on CMR should be interpreted in the context of HCM and in conjunction with other markers, such as LV hypertrophy or the presence of sarcomere mutation.
The clinical application of CMR tissue markers in HCM is currently limited to fibrosis detection. The distribution and severity of focal and interstitial fibrosis aid in differentiating HCM from its phenocopies, such as Fabry disease and amyloidosis [70]. Extensive replacement fibrosis is increasingly recognized as a prognostic marker for SCD and all-cause mortality [71], with LGE >15% of LV mass now serving as a second-line indication for implantable cardioverter-defibrillators [17, 72]. T1 mapping assists in distinguishing HCM from hypertensive LV hypertrophy [73], while ECV has been associated with heart failure outcomes [74]. An ongoing large observational study (NCT01915615), integrating genetic, blood, and CMR markers - including LGE, T1 mapping, and ECV - is likely to offer further insight into their prognostic capabilities in HCM [75]. CMR-based assessment of MVD and myocardial disarray remains research-focused and is not yet part of the etiological diagnosis of HCM. However, to date, cDTI, although in need of technical improvements [66], has demonstrated the ability to discriminate preclinical HCM from healthy controls [67], and its correlation with ventricular arrhythmias highlights its prognostic potential alongside LGE and ECV [65].
CMR tissue characterization techniques as a research tool in HCM
CMR, with its ability to accurately track pathological processes in the myocardium, is increasingly used in clinical trials of new therapeutics for non-ischemic cardiomyopathies [76]. CMR imaging series serve as surrogate markers of treatment efficacy and, as a merit, provide mechanistic insights into the molecular pathways of natural (placebo group) and treatment response over shorter time periods. In HCM, cardiac myosin inhibitors have been evaluated in clinical trials using surrogate clinical and imaging endpoints. The EXPLORER-HCM CMR substudy showed significant reductions in LV mass, wall thickness, and left atrial volume index, suggesting that mavacamten alters HCM pathophysiology [20]. Meanwhile, LGE and ECV were not significantly changed, supporting the irreversible nature of myocardial fibrosis. Recent research by Joy et al. showed associations of CMR-derived disarray and MVD with stages of phenotype evolution [67], suggesting potential future applications of these techniques in HCM research before and after therapeutic interventions. Notably, in overt disease, the presence versus absence of sarcomeric mutation has different effects on microstructure and microvasculature [67]. Stress perfusion CMR has recently been used as a validation tool for another potentially more cost-effective and clinically practical marker of MVD – impaired myocardial work on echocardiogram [77].
CMR techniques for discovery of circulating biomarkers in HCM
Several conventional blood biomarkers have been shown in association with CMR-derived tissue characteristics, particularly LGE [15]. NT-proBNP and troponin T exhibit positive correlations with increasing LGE and ECV levels in a graded manner [78, 79]. Other biomarkers linked to necrosis (troponin I), fibrosis (matrix metalloproteinase-9, endostatin, apelin), inflammation and apoptosis (high sensitivity C-reactive protein, TNF-alpha), and endothelial dysfunction (big endothelin-1) show correlations with LGE and MVD [15], although validation in larger studies is required.
Proteomic and transcriptomic studies aimed at identifying biomarker signatures associated with adverse myocardial changes observed on CMR are a relatively new line of research. However, several studies have already been conducted in various conditions, including HCM.
A large study in healthy individuals identified a circulating protein signature associated with interstitial fibrosis. Prospective follow-up using progression to heart failure as an endpoint may provide validation for the discovered protein panel [80]. In patients with heart failure and preserved LV ejection fraction, unique biomarker patterns correlated with ECV (7 proteins) and myocardial perfusion reserve (6 proteins) [81]. In HCM, quantitative proteomics identified a six-biomarker panel related to myocardial substrate changes and SCD risk, with five of the six biomarkers elevated in subclinical HCM patients [24]. Proteomic profiling of 701 patients with sarcomeric HCM identified circulating biomarkers associated with adverse imaging and clinical phenotypes, including LGE [82].
A complementary CMR and transcriptome profiling approach identified a circulating miR signature as a biomarker for LGE-positive cardiomyopathy in muscular dystrophy [83]. In HCM, the relationship between miRs and tissue CMR parameters has been investigated in six studies, all of which had relatively small sample sizes and primarily focused on fibrosis, employing LGE and T1-mapping techniques [48, 84–88] (Table 1). Several candidate individual miRs with moderate to strong diagnostic value have been identified. Notably, the study by Fang et al. demonstrated the significant superiority of miR panels over single miR markers for diagnostic purposes. In that study, individual miRs showed moderate diagnostic performance for interstitial fibrosis (AUC 0.663–0.742), while combining eight miRs substantially improved the diagnostic accuracy (AUC 0.87) [84].
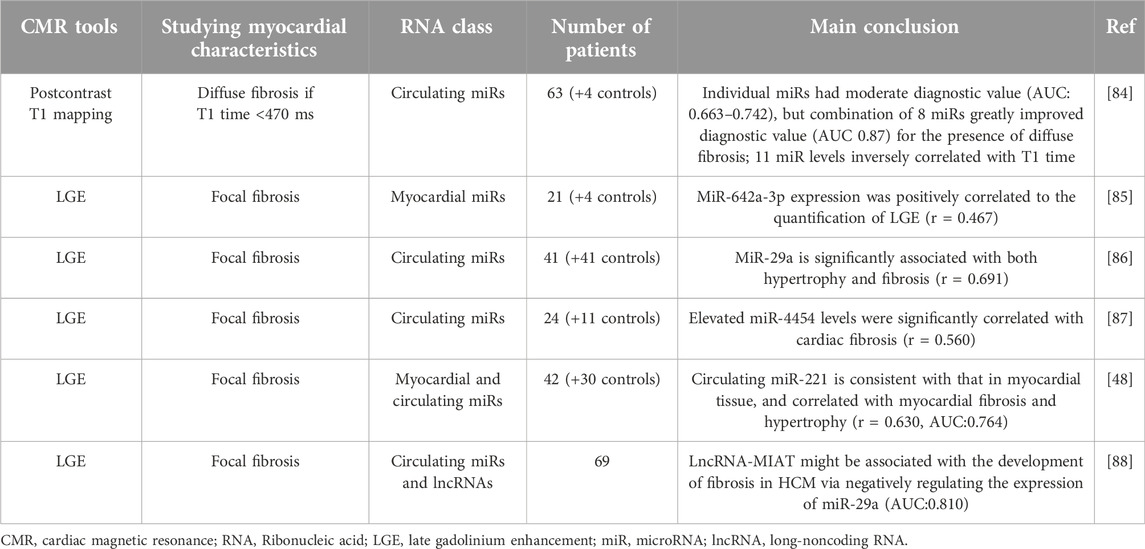
Table 1. Studies aimed at identifying MiR biomarkers associated with adverse myocardial changes observed on CMR in hypertrophic cardiomyopathy.
Discussion
MiRs are promising biomarkers, as their altering profiles can reflect distinct molecular processes in the heart. Given the heterogeneity of HCM, likely due to a cascade of molecular and structural changes, it is essential to investigate biomarkers across all stages of the disease. Before the onset of overt LV hypertrophy, HCM is characterized by abnormalities in myocardial microstructure, which emphasize their relevance in early pathogenesis. We presume that circulating molecules associated with these specific changes could be the strong candidates for further validation in larger studies as both disease-specific and prognostic biomarkers.
Given the limitations and biases of myocardial biopsy, CMR serves as a valuable tool for cardiac research. Advanced CMR technologies, currently being validated in preclinical models and human histology, enable the non-invasive visualization and quantification of myocardial microanatomical changes at all stages of the disease, including the preclinical phase. Preclinical HCM appears to be of particular interest for biomarker discovery, offering a unique opportunity to explore ncRNA signatures and potentially uncover disease-specific pathways without the confounding influence of secondary changes related to hemodynamic abnormalities. The expanding availability of genetic testing, including cascade family screening, is facilitating the identification of mutation carriers, making such studies feasible. When conducting discovery studies in overt HCM, the genotype of patients should be considered, as both miRs and myocardial microstructure are sensitive to the genotype status.
Novel HCM therapeutics, such as myosin inhibitors and gene editing, hold the potential to reverse the disease phenotype. Incorporating candidate miR panels into such self-controlled trials could enhance biomarker discovery by identifying those that reflect reversible adverse myocardial changes. A complementary imaging and genome-based biomarker approach could deeper insights into the complex underlying processes and identify novel targets for emerging therapeutic technologies.
HCM is a slowly progressive disease with a low event rate, making prospective studies is costly and time-consuming. Nevertheless, longitudinal studies with adequate sample size are essential to evaluate the prognostic power of candidate miR panels. Although conducting such studies in HCM is challenging, growing awareness and diagnostic expertise among professionals, emerging international cardiomyopathy collaborations, and advances in potentially curative therapies foster optimism.
Besides miRs, there are two other types of ncRNAs in the scope of interest in HCM: long non-coding RNAs (lncRNAs), which account for 80%–90% of all ncRNAs, and circular RNAs (circRNAs), a newer class of ncRNAs known for their stability due to a closed ring structure [89, 90]. To the best of our knowledge, only one study has investigated myocardial fibrosis in HCM by integrating CMR with relevant circulating lncRNAs [88]. This field remains largely unexplored, and significant efforts are required to advance our understanding.
Conclusion
Enhanced myocardial characterization and staging of HCM using advanced CMR techniques holds promise for identifying circulating miRs as biomarkers. MiR signatures associated with adverse microanatomical changes detected by CMR could be the strong candidates for longitudinal validation studies. To ensure comprehensive and reliable results, future research should consider patients’ genetic status.
Author contributions
OC proposed the concept, wrote the first draft of the manuscript and supervised the project. All authors contributed to the article and approved the submitted version.
Funding
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This research was supported by the Russian Science Foundation (grant No. 20-15-00353).
Acknowledgments
The authors would like to express their gratitude to Prof. Olga Favorova for her invaluable assistance in proofreading and editing the manuscript.
Conflict of interest
The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
References
1. Elliott, PM, Anastasakis, A, Borger, MA, Borggrefe, M, Cecchi, F, Charron, P, et al. 2014 ESC guidelines on diagnosis and management of hypertrophic cardiomyopathy: the task force for the diagnosis and management of hypertrophic cardiomyopathy of the European society of cardiology (ESC). Eur Heart J (2014) 35:2733–79. doi:10.1093/eurheartj/ehu284
2. Basso, C, Thiene, G, Corrado, D, Buja, G, Melacini, P, and Nava, A. Hypertrophic cardiomyopathy and sudden death in the young: pathologic evidence of myocardial ischemia. Hum Pathol (2000) 31(8):988–98. doi:10.1053/hupa.2000.16659
3. Maron, BJ, and Roberts, WC. Hypertrophic cardiomyopathy and cardiac muscle cell disorganization revisited: relation between the two and significance. Am Heart J (1981) 102(1):95–110. doi:10.1016/0002-8703(81)90419-1
4. Maron, BJ, Wolfson, JK, Epstein, SE, and Roberts, WC. Intramural (“small vessel”) coronary artery disease in hypertrophic cardiomyopathy. J Am Coll Cardiol (1986) 8(3):545–57. doi:10.1016/S0735-1097(86)80181-4
5. Varnava, AM, Elliott, PM, Mahon, N, Davies, MJ, and McKenna, WJ. Relation between myocyte disarray and outcome in hypertrophic cardiomyopathy. Am J Cardiol (2001) 88(3):275–9. doi:10.1016/S0002-9149(01)01640-X
6. Galati, G, Leone, O, Pasquale, F, Olivotto, I, Biagini, E, Grigioni, F, et al. Histological and histometric characterization of myocardial fibrosis in end-stage hypertrophic cardiomyopathy: a clinical-pathological study of 30 explanted hearts. Circ Heart Fail (2016) 9(9):e003090. doi:10.1161/CIRCHEARTFAILURE.116.003090
7. Soler, R, Méndez, C, Rodríguez, E, Barriales, R, Ochoa, JP, and Monserrat, L. Phenotypes of hypertrophic cardiomyopathy. An illustrative review of MRI findings. Insights Imaging (2018) 9(6):1007–20. doi:10.1007/s13244-018-0656-8
8. Marian, AJ, and Braunwald, E. Hypertrophic cardiomyopathy: genetics, pathogenesis, clinical manifestations, diagnosis, and therapy. Circ Res (2017) 121(7):749–70. doi:10.1161/CIRCRESAHA.117.311059
9. Ho, CY, Day, SM, Ashley, EA, Michels, M, Pereira, AC, Jacoby, D, et al. Genotype and lifetime burden of disease in hypertrophic cardiomyopathy: insights from the sarcomeric human cardiomyopathy registry (SHaRe). Circulation (2018) 138(14):1387–98. doi:10.1161/CIRCULATIONAHA.117.033200
10. Geisterfer-Lowrance, AA, Kass, S, Tanigawa, G, Vosberg, HP, McKenna, W, Seidman, CE, et al. A molecular basis for familial hypertrophic cardiomyopathy: a β cardiac myosin heavy chain gene missense mutation. Cell (1990) 62(5):999–1006. doi:10.1016/0092-8674(90)90274-I
11. Seidman, CE, and Seidman, JG. Identifying sarcomere gene mutations in hypertrophic cardiomyopathy: a personal history. Circ Res, 108(6)(2011). p. 743–50. doi:10.1161/CIRCRESAHA.110.223834
12. Spudich, JA. Three perspectives on the molecular basis of hypercontractility caused by hypertrophic cardiomyopathy mutations. Pflügers Archiv - Eur J Physiol (2019) 471(5):701–17. doi:10.1007/s00424-019-02259-2
13. Olivotto, I, Oreziak, A, Barriales-Villa, R, Abraham, TP, Masri, A, Garcia-Pavia, P, et al. Mavacamten for treatment of symptomatic obstructive hypertrophic cardiomyopathy (EXPLORER-HCM): a randomised, double-blind, placebo-controlled, phase 3 trial. Lancet (2020) 396(10253):759–69. doi:10.1016/S0140-6736(20)31792-X
14. McKenna, WJ, Crean, A, Greenway, S, Tadros, R, Veselka, J, and Woo, A. Hypertrophic cardiomyopathy: evolution to the present, ongoing challenges, and opportunities. Can J Cardiol (2024) 40(5):738–41. doi:10.1016/j.cjca.2024.03.005
15. Matthia, EL, Setteducato, ML, Elzeneini, M, Vernace, N, Salerno, M, Kramer, CM, et al. Circulating biomarkers in hypertrophic cardiomyopathy. J Am Heart Assoc (2022) 11(23):e027618. doi:10.1161/JAHA.122.027618
16. Ho, JE, Shi, L, Day, SM, Colan, SD, Russell, MW, Towbin, JA, et al. Biomarkers of cardiovascular stress and fibrosis in preclinical hypertrophic cardiomyopathy. Open Heart (2017) 4(2):e000615. doi:10.1136/openhrt-2017-000615
17. Ommen, SR, Ho, CY, Asif, IM, Balaji, S, Burke, MA, Day, SM, et al. 2024 AHA/ACC/AMSSM/HRS/PACES/SCMR guideline for the management of hypertrophic cardiomyopathy. J Am Coll Cardiol (2024) 83(23):2324–405. doi:10.1016/j.jacc.2024.02.014
18. O’Mahony, C, Jichi, F, Pavlou, M, Monserrat, L, Anastasakis, A, Rapezzi, C, et al. A novel clinical risk prediction model for sudden cardiac death in hypertrophic cardiomyopathy (HCM Risk-SCD). Eur Heart J (2014) 35(30):2010–20. doi:10.1093/eurheartj/eht439
19. Maron, MS, Masri, A, Choudhury, L, Olivotto, I, Saberi, S, Wang, A, et al. Phase 2 study of Aficamten in patients with obstructive hypertrophic cardiomyopathy. J Am Coll Cardiol (2023) 81(1):34–45. doi:10.1016/j.jacc.2022.10.020
20. Saberi, S, Cardim, N, Yamani, M, Schulz-Menger, J, Li, W, Florea, V, et al. Mavacamten favorably impacts cardiac structure in obstructive hypertrophic cardiomyopathy: EXPLORER-HCM cardiac magnetic resonance substudy analysis. Circulation (2021) 143(6):606–8. doi:10.1161/CIRCULATIONAHA.120.052359
21. Shimada, YJ, Raita, Y, Liang, LW, Maurer, MS, Hasegawa, K, Fifer, MA, et al. Comprehensive proteomics profiling reveals circulating biomarkers of hypertrophic cardiomyopathy. Circ Heart Fail (2021) 14(7):e007849. doi:10.1161/CIRCHEARTFAILURE.120.007849
22. Liang, LW, Hasegawa, K, Maurer, MS, Reilly, MP, Fifer, MA, and Shimada, YJ. Comprehensive transcriptomics profiling of MicroRNA reveals plasma circulating biomarkers of hypertrophic cardiomyopathy and dysregulated signaling pathways. Circ Heart Fail (2023) 16(6):e010010. doi:10.1161/CIRCHEARTFAILURE.122.010010
23. Sucharov, CC, Neltner, B, Pietra, AE, Karimpour-Fard, A, Patel, J, Ho, CY, et al. Circulating MicroRNAs identify early phenotypic changes in sarcomeric hypertrophic cardiomyopathy. Circ Heart Fail (2023) 16(6):e010291. doi:10.1161/CIRCHEARTFAILURE.122.010291
24. Captur, G, Heywood, WE, Coats, C, Rosmini, S, Patel, V, Lopes, LR, et al. Identification of a multiplex biomarker panel for hypertrophic cardiomyopathy using quantitative proteomics and machine learning. Mol and Cell Proteomics (2020) 19(1):114–27. doi:10.1074/mcp.RA119.001586
25. Small, EM, and Olson, EN. Pervasive roles of microRNAs in cardiovascular biology. Nature (2011) 469(7330):336–42. doi:10.1038/nature09783
26. Misir, S, Wu, N, and Yang, BB. Specific expression and functions of circular RNAs. Cell Death Differ (2022) 29(3):481–91. doi:10.1038/s41418-022-00948-7
27. Yao, RW, Wang, Y, and Chen, LL. Cellular functions of long noncoding RNAs. Nat Cel Biol (2019) 21(5):542–51. doi:10.1038/s41556-019-0311-8
28. Ambros, V. The functions of animal microRNAs. Nature (2004) 431(7006):350–5. doi:10.1038/nature02871
29. Zhao, Y, Wang, Z, Zhang, W, and Zhang, L. MicroRNAs play an essential role in autophagy regulation in various disease phenotypes. BioFactors (2019) 45(6):844–56. doi:10.1002/biof.1555
30. Das, A, Samidurai, A, and Salloum, FN. Deciphering non-coding RNAs in cardiovascular health and disease. Front Cardiovasc Med (2018) 5:73. doi:10.3389/fcvm.2018.00073
31. Bauersachs, J, Solomon, SD, Anker, SD, Antorrena-Miranda, I, Batkai, S, Viereck, J, et al. Efficacy and safety of CDR132L in patients with reduced left ventricular ejection fraction after myocardial infarction: rationale and design of the HF-REVERT trial. Eur J Heart Fail (2024) 26(3):674–82. doi:10.1002/ejhf.3139
32. Seal, RL, Chen, L, Griffiths-Jones, S, Lowe, TM, Mathews, MB, O’Reilly, D, et al. A guide to naming human non-coding RNA genes. EMBO J (2020) 39(6):e103777. doi:10.15252/embj.2019103777
33. Kozomara, A, Birgaoanu, M, and Griffiths-Jones, S. miRBase: from microRNA sequences to function. Nucleic Acids Res (2019) 47(D1):D155–D162. doi:10.1093/nar/gky1141
34. Ntelios, D, Georgiou, E, Alexouda, S, Malousi, A, Efthimiadis, G, and Tzimagiorgis, G. A critical approach for successful use of circulating microRNAs as biomarkers in cardiovascular diseases: the case of hypertrophic cardiomyopathy. Heart Fail Rev (2022) 27(1):281–94. doi:10.1007/s10741-021-10084-y
36. Mitchell, PS, Parkin, RK, Kroh, EM, Fritz, BR, Wyman, SK, Pogosova-Agadjanyan, EL, et al. Circulating microRNAs as stable blood-based markers for cancer detection. Proc Natl Acad Sci USA (2008) 105(30):10513–8. doi:10.1073/pnas.0804549105
37. Vickers, KC, Palmisano, BT, Shoucri, BM, Shamburek, RD, and Remaley, AT. MicroRNAs are transported in plasma and delivered to recipient cells by high-density lipoproteins. Nat Cel Biol (2011) 13(4):423–33. doi:10.1038/ncb2210
38. Margolis, L, and Sadovsky, Y. The biology of extracellular vesicles: the known unknowns. Plos Biol (2019) 17(7):e3000363. doi:10.1371/journal.pbio.3000363
39. Bagnall, RD, Tsoutsman, T, Shephard, RE, Ritchie, W, and Semsarian, C. Global MicroRNA profiling of the mouse ventricles during development of severe hypertrophic cardiomyopathy and heart failure. PLoS One (2012) 7:9e44744. doi:10.1371/journal.pone.0044744
40. Guo, M, Luo, J, Zhao, J, Shang, D, Lv, Q, and Zang, P. Combined use of circulating miR-133a and NT-proBNP improves heart failure diagnostic accuracy in elderly patients. Med Sci Monit (2018) 24:8840–8. doi:10.12659/MSM.911632
41. Wong, LL, Zou, R, Zhou, L, Lim, JY, Phua, DCY, Liu, C, et al. Combining circulating MicroRNA and NT-proBNP to detect and categorize heart failure subtypes. J Am Coll Cardiol (2019) 73(11):1300–13. doi:10.1016/j.jacc.2018.11.060
42. Carè, A, Catalucci, D, Felicetti, F, Bonci, D, Addario, A, Gallo, P, et al. MicroRNA-133 controls cardiac hypertrophy. Nat Med (2007) 13(5):613–8. doi:10.1038/nm1582
43. Leptidis, S, El Azzouzi, H, Lok, SI, de Weger, R, Olieslagers, S, Kisters, N, et al. A deep sequencing approach to uncover the miRNOME in the human heart. PLoS One (2013) 8(2):e57800. doi:10.1371/journal.pone.0057800
44. Kuster, DWD, Mulders, J, Ten Cate, FJ, Michels, M, Dos Remedios, CG, da Costa Martins, PA, et al. MicroRNA transcriptome profiling in cardiac tissue of hypertrophic cardiomyopathy patients with MYBPC3 mutations. J Mol Cell Cardiol (2013) 65:59–66. doi:10.1016/j.yjmcc.2013.09.012
45. Song, L, Su, M, Wang, S, Zou, Y, Wang, X, Wang, Y, et al. MiR-451 is decreased in hypertrophic cardiomyopathy and regulates autophagy by targeting TSC 1. J Cell Mol Med (2014) 18(11):2266–74. doi:10.1111/jcmm.12380
46. Gao, J, Collyer, J, Wang, M, Sun, F, and Xu, F. Genetic dissection of hypertrophic cardiomyopathy with myocardial RNA-seq. Int J Mol Sci (2020) 21(9):3040. doi:10.3390/ijms21093040
47. Palacín, M, Reguero, JR, Martín, M, Díaz Molina, B, Morís, C, Alvarez, V, et al. Profile of MicroRNAs differentially produced in hearts from patients with hypertrophic cardiomyopathy and sarcomeric mutations. Clin Chem (2011) 57(11):1614–6. doi:10.1373/clinchem.2011.168005
48. Huang, D, Chen, Z, Wang, J, Chen, Y, Liu, D, and Lin, K. MicroRNA-221 is a potential biomarker of myocardial hypertrophy and fibrosis in hypertrophic obstructive cardiomyopathy. Biosci Rep (2020) 40(1):BSR20191234. doi:10.1042/BSR20191234
49. Baulina, N, Pisklova, M, Kiselev, I, Chumakova, O, Zateyshchikov, D, and Favorova, O. Circulating miR-499a-5p is a potential biomarker of MYH7—associated hypertrophic cardiomyopathy. Int J Mol Sci (2022) 23(7):3791. doi:10.3390/ijms23073791
50. Van Rooij, E, Quiat, D, Johnson, BA, Sutherland, LB, Qi, X, Richardson, JA, et al. A family of microRNAs encoded by myosin genes governs myosin expression and muscle performance. Develop Cel (2009) 17(5):662–73. doi:10.1016/j.devcel.2009.10.013
51. Sood, P, Krek, A, Zavolan, M, Macino, G, and Rajewsky, N. Cell-type-specific signatures of microRNAs on target mRNA expression. Proc Natl Acad Sci (2006) 103(8):2746–51. doi:10.1073/pnas.0511045103
52. Chistiakov, DA, Orekhov, AN, and Bobryshev, YV. Cardiac-specific miRNA in cardiogenesis, heart function, and cardiac pathology (with focus on myocardial infarction). J Mol Cell Cardiol (2016) 94:107–21. doi:10.1016/j.yjmcc.2016.03.015
53. Liu, N, and Olson, EN. MicroRNA regulatory networks in cardiovascular development. Develop Cel (2010) 18(4):510–25. doi:10.1016/j.devcel.2010.03.010
54. Düsing, P, Zietzer, A, and Jansen, F. MicroRNA-based diagnostics in heart diseases. JACC: Basic Translational Sci (2021) 6(11):897–9. doi:10.1016/j.jacbts.2021.08.004
55. De Gaspari, M, Basso, C, Perazzolo Marra, M, Elia, S, Bueno Marinas, M, Angelini, A, et al. Small vessel disease: another component of the hypertrophic cardiomyopathy phenotype not necessarily associated with fibrosis. J Clin Med (2021) 10(4):575. doi:10.3390/jcm10040575
56. Garcia-Canadilla, P, Cook, AC, Mohun, TJ, Oji, O, Schlossarek, S, Carrier, L, et al. Myoarchitectural disarray of hypertrophic cardiomyopathy begins pre-birth. J Anat (2019) 235(5):962–76. doi:10.1111/joa.13058
57. McKenna, WJ, Stewart, JT, Nihoyannopoulos, P, McGinty, F, and Davies, MJ. Hypertrophic cardiomyopathy without hypertrophy: two families with myocardial disarray in the absence of increased myocardial mass. Heart (1990) 63(5):287–90. doi:10.1136/hrt.63.5.287
58. Varnava, A, Baboonian, C, Davison, F, de Cruz, L, Elliott, PM, Davies, MJ, et al. A new mutation of the cardiac troponin T gene causing familial hypertrophic cardiomyopathy without left ventricular hypertrophy. Heart (1999) 82(5):621–4. doi:10.1136/hrt.82.5.621
59. Green, EM, Wakimoto, H, Anderson, RL, Evanchik, MJ, Gorham, JM, Harrison, BC, et al. A small-molecule inhibitor of sarcomere contractility suppresses hypertrophic cardiomyopathy in mice. Science (2016) 351(6273):617–21. doi:10.1126/science.aad3456
60. Messroghli, DR, Moon, JC, Ferreira, VM, Grosse-Wortmann, L, He, T, Kellman, P, et al. Clinical recommendations for cardiovascular magnetic resonance mapping of T1, T2, T2* and extracellular volume: a consensus statement by the Society for Cardiovascular Magnetic Resonance (SCMR) endorsed by the European Association for Cardiovascular Imaging (EACVI). J Cardiovasc Magn Reson (2016) 19(1):75. doi:10.1186/s12968-017-0389-8
61. Miller, CA, Naish, JH, Bishop, P, Coutts, G, Clark, D, Zhao, S, et al. Comprehensive validation of cardiovascular magnetic resonance techniques for the assessment of myocardial extracellular volume. Circ Cardiovasc Imaging (2013) 6(3):373–83. doi:10.1161/CIRCIMAGING.112.000192
62. Schelbert, EB, Hsu, LY, Anderson, SA, Mohanty, BD, Karim, SM, Kellman, P, et al. Late gadolinium-enhancement cardiac magnetic resonance identifies postinfarction myocardial fibrosis and the border zone at the near cellular level in ex vivo rat heart. Circ Cardiovasc Imaging (2010) 3(6):743–52. doi:10.1161/CIRCIMAGING.108.835793
63. Iles, LM, Ellims, AH, Llewellyn, H, Hare, JL, Kaye, DM, McLean, CA, et al. Histological validation of cardiac magnetic resonance analysis of regional and diffuse interstitial myocardial fibrosis. Eur Heart J - Cardiovasc Imaging (2015) 16(1):14–22. doi:10.1093/ehjci/jeu182
64. Moon, JCC, Reed, E, Sheppard, MN, Elkington, AG, Ho, SY, Burke, M, et al. The histologic basis of late gadolinium enhancement cardiovascular magnetic resonance in hypertrophic cardiomyopathy. J Am Coll Cardiol (2004) 43(12):2260–4. doi:10.1016/j.jacc.2004.03.035
65. Ariga, R, Tunnicliffe, EM, Manohar, SG, Mahmod, M, Raman, B, Piechnik, SK, et al. Identification of myocardial disarray in patients with hypertrophic cardiomyopathy and ventricular arrhythmias. J Am Coll Cardiol (2019) 73(20):2493–502. doi:10.1016/j.jacc.2019.02.065
66. Dall'Armellina, E, Ennis, DB, Axel, L, Croisille, P, Ferreira, PF, Gotschy, A, et al. Cardiac diffusion-weighted and tensor imaging: a Society for Cardiovascular Magnetic Resonance (SCMR) special interest group consensus statement. J Cardiovasc Magn Reson (2024) 21:101109. doi:10.1016/j.jocmr.2024.101109
67. Joy, G, Kelly, CI, Webber, M, Pierce, I, Teh, I, McGrath, L, et al. Microstructural and microvascular phenotype of sarcomere mutation carriers and overt hypertrophic cardiomyopathy. Circulation (2023) 148(10):808–18. doi:10.1161/CIRCULATIONAHA.123.063835
68. Petersen, SE, Jerosch-Herold, M, Hudsmith, LE, Robson, MD, Francis, JM, Doll, HA, et al. Evidence for microvascular dysfunction in hypertrophic cardiomyopathy: new insights from multiparametric magnetic resonance imaging. Circulation (2007) 115(18):2418–25. doi:10.1161/CIRCULATIONAHA.106.657023
69. Villa, AD, Sammut, E, Zarinabad, N, Carr-White, G, Lee, J, Bettencourt, N, et al. Microvascular ischemia in hypertrophic cardiomyopathy: new insights from high-resolution combined quantification of perfusion and late gadolinium enhancement. J Cardiovasc Magn Reson (2016) 18:4. doi:10.1186/s12968-016-0223-8
70. Arbelo, E, Protonotarios, A, Gimeno, JR, Arbustini, E, Barriales-Villa, R, Basso, C, et al. 2023 ESC Guidelines for the management of cardiomyopathies. Eur Heart J (2023) 44(37):3503–626. doi:10.1093/eurheartj/ehad194
71. Kamp, NJ, Chery, G, Kosinski, AS, Desai, MY, Wazni, O, Schmidler, GS, et al. Risk stratification using late gadolinium enhancement on cardiac magnetic resonance imaging in patients with hypertrophic cardiomyopathy: a systematic review and meta-analysis. Prog Cardiovasc Dis (2021) 66:10–6. doi:10.1016/j.pcad.2020.11.001
72. Zeppenfeld, K, Tfelt-Hansen, J, de Riva, M, Winkel, BG, Behr, ER, Blom, NA, et al. 2022 ESC Guidelines for the management of patients with ventricular arrhythmias and the prevention of sudden cardiac death. Eur Heart J (2022) 43(40):3997–4126. doi:10.1093/eurheartj/ehac262
73. Hinojar, R, Varma, N, Child, N, Goodman, B, Jabbour, A, Yu, CY, et al. T1 mapping in discrimination of hypertrophic phenotypes: hypertensive heart disease and hypertrophic cardiomyopathy: findings from the international T1 multicenter cardiovascular magnetic resonance study. Circ Cardiovasc Imaging (2015) 8(12):e003285. doi:10.1161/CIRCIMAGING.115.003285
74. Yang, EY, Ghosn, MG, Khan, MA, Gramze, NL, Brunner, G, Nabi, F, et al. Myocardial extracellular volume fraction adds prognostic information beyond myocardial replacement fibrosis. Circ Cardiovasc Imaging (2019) 12(12):e009535. doi:10.1161/CIRCIMAGING.119.009535
75. Kramer, CM, Appelbaum, E, Desai, MY, Desvigne-Nickens, P, DiMarco, JP, Friedrich, MG, et al. Hypertrophic Cardiomyopathy Registry: the rationale and design of an international, observational study of hypertrophic cardiomyopathy. Am Heart J (2015) 170(2):223–30. doi:10.1016/j.ahj.2015.05.013
76. Benz, DC, Gräni, C, Antiochos, P, Heydari, B, Gissler, MC, Ge, Y, et al. Cardiac magnetic resonance biomarkers as surrogate endpoints in cardiovascular trials for myocardial diseases. Eur Heart J (2023) 44(45):4738–47. doi:10.1093/eurheartj/ehad510
77. Garcia Brás, P, Rosa, SA, Cardoso, I, Branco, LM, Galrinho, A, Gonçalves, AV, et al. Microvascular dysfunction is associated with impaired myocardial work in obstructive and nonobstructive hypertrophic cardiomyopathy: a multimodality study. J Am Heart Assoc (2023) 12(8):e028857. doi:10.1161/JAHA.122.028857
78. Neubauer, S, Kolm, P, Ho, CY, Kwong, RY, Desai, MY, Dolman, SF, et al. Distinct subgroups in hypertrophic cardiomyopathy in the NHLBI HCM registry. J Am Coll Cardiol (2019) 74(19):2333–45. doi:10.1016/j.jacc.2019.08.1057
79. Ho, CY, Abbasi, SA, Neilan, TG, Shah, RV, Chen, Y, Heydari, B, et al. T1 measurements identify extracellular volume expansion in hypertrophic cardiomyopathy sarcomere mutation carriers with and without left ventricular hypertrophy. Circ Cardiovasc Imaging (2013) 6(3):415–22. doi:10.1161/CIRCIMAGING.112.000333
80. Bakhshi, H, Michelhaugh, SA, Bruce, SA, Seliger, SL, Qian, X, Ambale Venkatesh, B, et al. Association between proteomic biomarkers and myocardial fibrosis measured by MRI: the multi-ethnic study of atherosclerosis. EBioMedicine (2023) 90:104490. doi:10.1016/j.ebiom.2023.104490
81. Siggins, C, Pan, JA, Löffler, AI, Yang, Y, Shaw, PW, Balfour, PC, et al. Cardiometabolic biomarker patterns associated with cardiac MRI defined fibrosis and microvascular dysfunction in patients with heart failure with preserved ejection fraction. Front Cardiovasc Med (2024) 11:1334226. doi:10.3389/fcvm.2024.1334226
82. Tahir, UA, Kolm, P, Kwong, RY, Desai, MY, Dolman, SF, Deng, S, et al. Protein biomarkers of adverse clinical features and events in sarcomeric hypertrophic cardiomyopathy. Circ Heart Fail (2024) 5:e011707. doi:10.1161/CIRCHEARTFAILURE.124.011707
83. Becker, S, Florian, A, Patrascu, A, Rösch, S, Waltenberger, J, Sechtem, U, et al. Identification of cardiomyopathy associated circulating miRNA biomarkers in patients with muscular dystrophy using a complementary cardiovascular magnetic resonance and plasma profiling approach. J Cardiovasc Magn Reson (2016) 18(1):25. doi:10.1186/s12968-016-0244-3
84. Fang, L, Ellims, AH, Moore, XL, White, DA, Taylor, AJ, Chin-Dusting, J, et al. Circulating microRNAs as biomarkers for diffuse myocardial fibrosis in patients with hypertrophic cardiomyopathy. J Transl Med (2015) 13(1):314. doi:10.1186/s12967-015-0672-0
85. Zhang, C, Zhang, H, Zhao, L, Wei, Z, Lai, Y, and Ma, X. Differential expression of microRNAs in hypertrophied myocardium and their relationship to late gadolinium enhancement, left ventricular hypertrophy and remodeling in hypertrophic cardiomyopathy. Diagnostics (2022) 12(8):1978. doi:10.3390/diagnostics12081978
86. Roncarati, R, Viviani Anselmi, C, Losi, MA, Papa, L, Cavarretta, E, Da Costa Martins, P, et al. Circulating miR-29a, among other up-regulated MicroRNAs, is the only biomarker for both hypertrophy and fibrosis in patients with hypertrophic cardiomyopathy. J Am Coll Cardiol (2014) 63(9):920–7. doi:10.1016/j.jacc.2013.09.041
87. Thottakara, T, Lund, N, Krämer, E, Kirchhof, P, Carrier, L, and Patten, M. A novel miRNA screen identifies miRNA-4454 as a candidate biomarker for ventricular fibrosis in patients with hypertrophic cardiomyopathy. Biomolecules (2021) 11(11):1718. doi:10.3390/biom11111718
88. Zhou, J, Zhou, Y, and Wang, C. LncRNA-MIAT regulates fibrosis in hypertrophic cardiomyopathy (HCM) by mediating the expression of miR-29a-3p. J Cell Biochem (2019) 120(5):7265–75. doi:10.1002/jcb.28001
89. Baulina, NM, Kiselev, IS, Chumakova, OS, and Favorova, OO. Circular RNAs: biogenesis, functions, and role in myocardial hypertrophy. Biochemistry (Mosc) (2024) 89(S1):S1–S13. doi:10.1134/S0006297924140013
Keywords: hypertrophic cardiomyopathy, non-coding RNAs, microRNAs, cardiac magnetic resonance, biomarkers, fibrosis, disarray, microvasculature
Citation: Chumakova OS and Mershina EA (2024) Circulating microRNA as promising biomarkers in hypertrophic cardiomyopathy: can advanced cardiac magnetic resonance unlock new insights in research?. Exp. Biol. Med. 249:10334. doi: 10.3389/ebm.2024.10334
Received: 02 August 2024; Accepted: 28 November 2024;
Published: 18 December 2024.
Copyright © 2024 Chumakova and Mershina. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Olga S. Chumakova, Y2h1bWFrb3Zhb2xnYUBiay5ydQ==
 Olga S. Chumakova
Olga S. Chumakova Elena A. Mershina
Elena A. Mershina