- 1The School of Clinical Medicine, Fujian Medical University, Fuzhou, Fujian, China
- 2Department of Hematology, The First Affiliated Hospital of Xiamen University and Institute of Hematology, School of Medicine, Xiamen University, Xiamen, China
- 3Department of Hematology, Shantou Central Hospital, Shantou, Guangdong, China
Abstract
Acute myeloid leukemia (AML) is one of the most threatening hematological malignances. cGAS-STING pathway plays an important role in tumor immunity and development. However, the prognostic role of cGAS-STING pathway in AML remains unknown. Firstly, The expression of cGAS and STING was analyzed by bioinformatics analysis. Subsequently, Bone marrow samples were collected from 120 AML patients and 15 healthy individuals in an independent cohort. The cGAS and STING expression was significantly elevated in AML patients compared with healthy controls. Patients with high cGAS and STING expression had a higher NRAS/KRAS mutation rate and lower complete remission (CR) rate. High cGAS and STING expression was significantly associated with lower overall survival (OS) and disease-free survival (DFS). Our findings revealed that the expression levels of cGAS and STING in AML are elevated. High expression of cGAS and STING correlated with worse OS and DFS and may be a useful biomarker for inferior prognosis in AML patients.
Impact statement
Acute myeloid leukemia (AML) is an aggressive hematopoietic malignancy with a high incidence rate and poor clinical prognosis. However, the current understanding of the molecular mechanism of AML development and progress is very limited. In our study, we evaluated the expression of cGAS and STING by collecting bone marrow samples from 120 AML patients and 15 healthy individuals, and found that cGAS-STING pathway was involved in the pathogenesis of AML. The high expression of cGAS and STING is related to the worse OS and DFS, which may be useful biomarkers for the poor prognosis of AML patients. Our research fills the gap in the pathogenesis of AML and provides potential biomarkers for clinical diagnosis and treatment.
Introduction
Acute myeloid leukemia (AML) is an aggressive hematopoietic malignant disease resulted in high morbidity and unfavorable clinical outcome [1]. Cytogenetics is the backbone for risk stratification, facilitating the classification of AML patients into favorable, intermediate, and poor prognostic groups. However, more than half of AML patients are classified as an intermediate cytogenetic risk group but their clinical outcomes turn out to be distinct [2, 3]. A large percentage of AML patients will suffer from disease recurrences due to the heterogeneous AML clones [4, 5]. The current perception of molecular mechanisms on the development and progression of AML is limited to date [6]. Thus, the identification of new molecular biomarker and revealing of novel mechanism are needed to prompt a more precise risk stratification and develop targeted therapies for AML.
As a vital DNA sensor, cyclic guanosine monophos-phate (GMP)-adenosine monophosphate (AMP) (cGAMP) synthase (cGAS) initiated an innate immunity pathway through binding deviant DNA in the cytosol. cGAMP can activate stimulator of interferon genes (STING), leading to a signaling cascade which produces type I interferons and other functional cytokines [7]. cGAS-STING pathway was previously introduced as a crucial initiator of innate immune and anti-virus responses [8]. Recent studies have revealed the multiple role of cGAS-STING pathway in cancer. Activated cGAS-STING pathway in tumor cells lead to upregulation of various inflammatory genes, such as Type I interferon and impedes the neoplastic progression [9]. There are also related reports in AML, and therefore, many scientists have attempted to increase the expression of type I interferons by upregulating STING, thus achieving the goal of treating AML [10–13]. Yet, mounting evidence indicates that cGAS-STING pathway might provoke inflammation, leading to tumor transformation, development and metastasis in certain diseases [14–16]. Consequently, the relationship between the expression levels of cGAS and Sting in AML and patient prognosis remains unclear.
In this study, we found that cGAS and STING expression levels were higher in AML patients compared with healthy controls by using Gene Expression Profiling Interactive Analysis (GEPIA) and Gene-Set Enrichment Analysis (GSEA) with further validation performed in our cohort. Furthermore, we investigated the impact of cGAS and STING expression on the clinical outcomes of AML patients. Our results indicated that higher expression of cGAS and STING was associated with inferior survival in AML patients.
Materials and Methods
Datasets
The GEPIA1 integrated the two databases including The Cancer Genome Atlas (TCGA)2 and the Genotype-Tissue Expression Project (GTEx)3. This platform can perform gene expression analysis based on RNA-seq expression data for 9,736 tumor samples and 8,587 control samples [17]. In this study, the GEPIA was used to analyze the expression of cGAS and STING in different tumors. Two gene expression profile datasets (GSE63270 and GSE30029) which involved expression data for healthy and AML bone marrow samples were downloaded from Gene Expression Omnibus (GEO,4) [18].
Clinical patients and reverse transcribed quantitative PCR (RT-qPCR)
A total of 120 non-M3 AML patients diagnosed in our department during 2018–2020 were enrolled in the present study. Additionally, 15 healthy allo-HSCT donors were enrolled as control. All patients were diagnosed and classified according to French -American—British (FAB) group and World Health Organization (WHO) classification. Complete remission (CR) was defined by <5% blast cells in the bone marrow and normalization of the peripheral blood counts at 4 weeks after starting induction therapy, without any evidence of extramedullary disease. The written informed consents were provided from all patients in accordance with Declaration of Helsinki.
Ficoll-Hypaque density gradient column (Cytova, Uppsala, Sweden) was used to isolate monocytes. The total RNA was isolated from monocytes by Trizol (Invitrogen, United States), then reverse transcribed into cDNA using BioTeke super RT Kit (BioTeke, Beijing, China). RT-qPCR was performed with an ABI PRISM 7500 real-time PCR system (PE Applied Biosystems, Foster City, CA, United States). We selected β-actin as a control gene to compensate for variations in quality and quantity of RNA and cDNA. The amplification conditions were as follows: 95 °C for 2 min, followed by 40 cycles of 95 °C for 15 s and 60 °C for 30 s. Sequences were as follows:
cGAS - Forward: 5′- CACGAAGCCAAGACCTCCG -3′
cGAS - Reverse: 5′- GTCGCACTTCAGTCTGAGCA -3′
STING - Forward: 5′- CCAGAGCACACTCTCCGGTA -3′
STING - Reverse: 5′- CGCATTTGGGAGGGAGTAGTA -3′
β-actin - Forward: 5′- TGTGGCATCCACGAAACTAC -3′
β-actin - Reverse: 5′- GGAGCAATGATCTTGATCTTCA -3′
The relative expression levels of above genes were calculated using the 2−ΔΔCT method (fold change over control expression).
Statistical analysis
All data were analyzed with SPSS (version 20.0; Chicago, IL) and GraphPad Prism 5.0 (GraphPad Software Inc., United States). Overall survival (OS) was calculated from the date of diagnosis until death caused by any reason. Disease-free survival (DFS) was defined as the time from achievement of CR to relapse or the last follow-up. Mann–Whitney U test and Chi-square test were used for continuous and categorical variables respectively. The probabilities of OS and DFS were estimated using the Kaplan-Meier method. The expression of cGAS and STING as well as other variables were included in the univariate analysis. Only variables with p < 0.1 were included in a Cox proportional hazards model with time-dependent variables. Spear-man rank correlation was used to analyze the correlation between two variables. Unless otherwise specified, p values were based on two-sided hypothesis tests. Alpha was set at 0.05.
Results
Elevated expression levels of cGAS and STING in AML
Firstly, we analyzed the expression levels of cGAS and STING in different types of tumors using GEPIA database. The analysis revealed that cGAS and STING expression levels were higher in AML compared to other tumors (Figure 1A). In addition, the dataset from GEPIA which involved 173 AML and 70 healthy control samples showed that cGAS and STING gene expression was higher in AML samples than healthy controls (Figure 1B). We then analyzed available molecular data from GSE63270 (involved 62 AML and 42 normal controls) and GSE30029 (involved 46 AML and 31 normal controls). In both cohorts, cGAS and STING gene expression levels were higher in AML samples than normal controls in consistence with the result from GEPIA analysis (p < 0.0001, Figures 1C, D). To validate the results from above publicly available datasets, we collected bone marrow samples from 120 AML patients and 15 healthy donors and performed the detection of cGAS and STING gene expression. We found that the expression of cGAS and STING was also elevated in AML patients from our cohort (p < 0.0001, Figure 1E). Taken together, these results demonstrate that up-regulation of cGAS and STING is a common feature in AML.
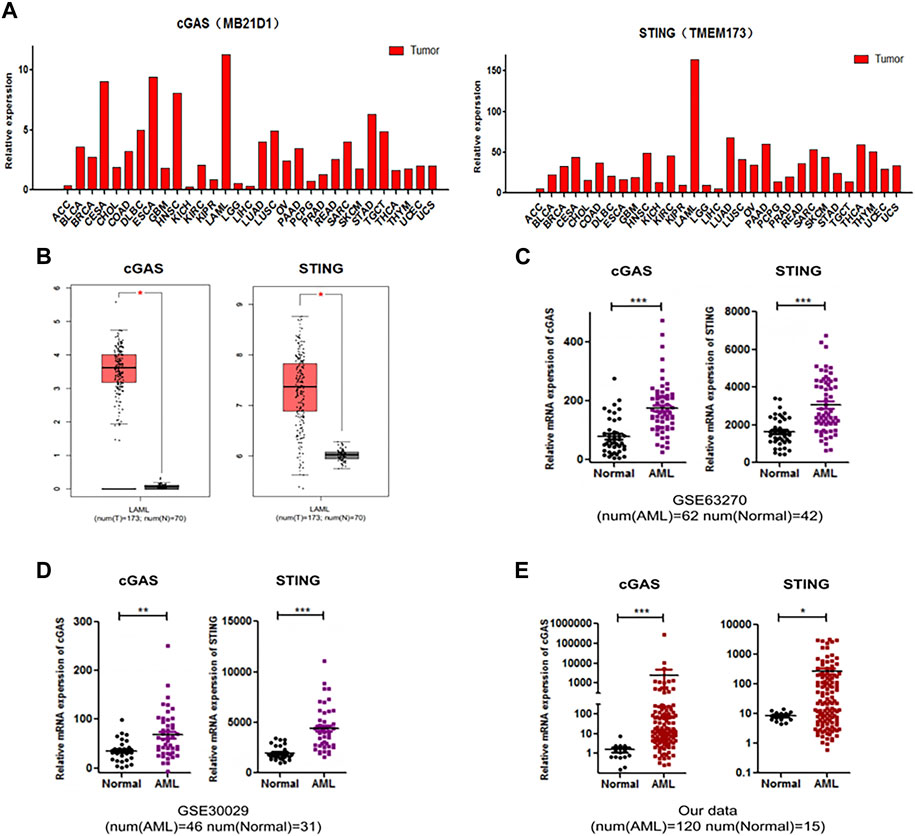
Figure 1. The expression levels of cGAS (MB21D1) and SING (TMEM173) were elevated in AML. (A) Analysis of the expression of cGAS and SING in different types of tumors, from GEPIA database. (B–D) Up-regulated cGAS and STING expression in AML patients compared with normal controls, from GEPIA database (173 AML and 70 normal controls), GEO: GSE63270 (62 AML and 42 normal controls) and GSE30029 (46 AML and 31 normal controls), respectively. (E) Validation of cGAS and STING expression of AML patients and normal controls in an independent cohort (120 AML and 15 normal controls). *p < 0.05; **p < 0.01; ***p < 0.001.
Patient characteristics
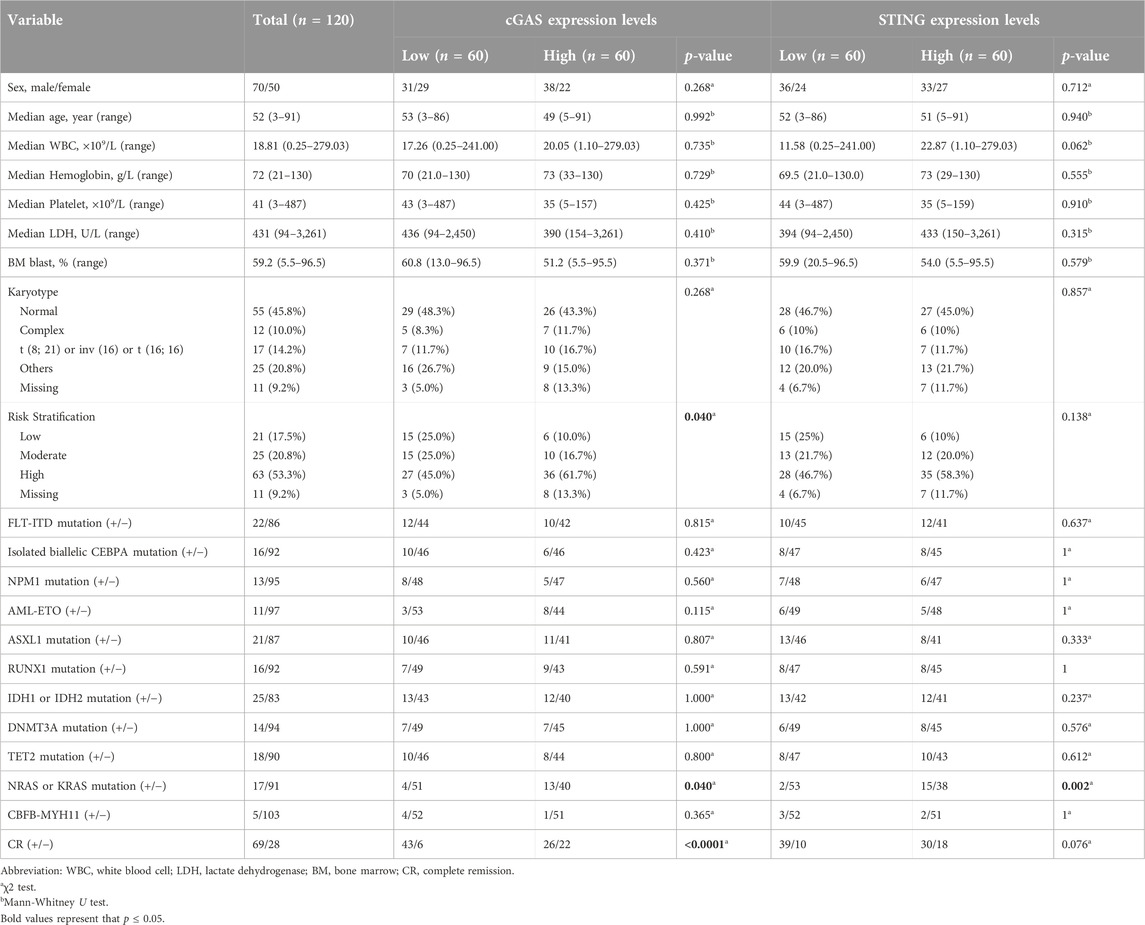
The baseline characteristics of patients are shown in Table 1. We dichotomized the patients into two high and low groups based on the median values of cGAS and STING expression respectively. The distribution of risk stratification was significantly different in cGAS high and low groups with more patients at high risk in cGAS high group (p = 0.040). Patients with higher cGAS expression had a higher NRAS/KRAS mutation rate (p = 0.040) and lower CR rate (p < 0.0001). Similarly, patients in STING high group had a higher NRAS/KRAS mutation rate (p = 0.002) and tended to have a lower CR rate (p = 0.076). The other characteristics including age, white blood count, hemoglobin, platelet, lactate dehydrogenase, bone marrow blast percentage, karyotypes, FLT3-ITD mutation, isolated biallelic CEBPA mutation, NPM1 mutation and other mutation between patients with high and low cGAS or STING expression were not significantly different.
High cGAS and STING expression correlated to inferior survival in AML
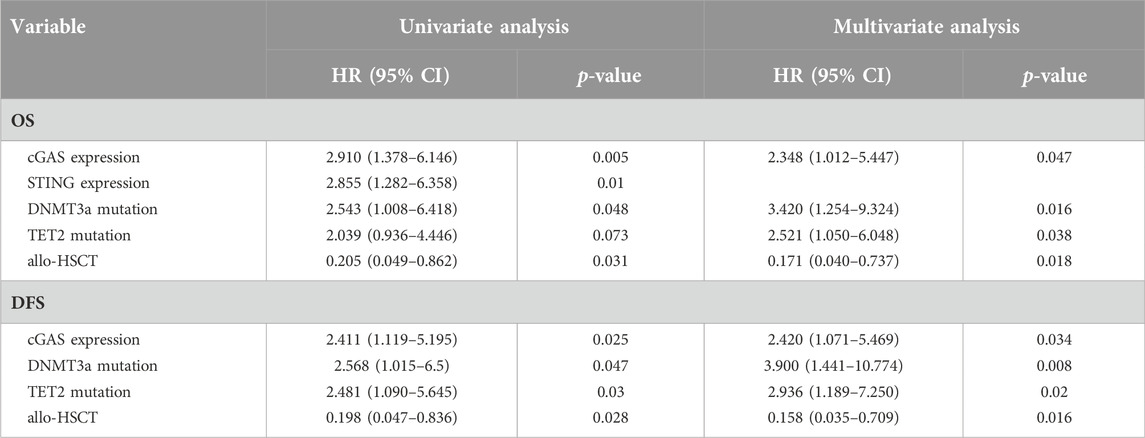
We further analyzed the overall survival (OS) in an adjusted cohort which excluded 18 untreated patients, 18 patients received allo-HSCT and 5 patients lost to follow-up. The remaining patients received similar treatments. In analysis of disease-free survival (DFS), 6 patients who didn’t achieve CR after treatment were further excluded. Kaplan-Meier analysis showed that patients with higher cGAS expression had a shorter OS (377.6 vs. 626.7 days, p = 0.007, Figure 2A) as well as a shorter DFS (312.2 vs. 543.9 days, p = 0.012, Figure 2B). Likewise, patients with higher STING expression had a shorter OS (332.9 vs. 612.7 days, p = 0.004, Figure 2C) as well as a shorter DFS (178.3 vs. 507.8 days, p = 0.034, Figure 2D).
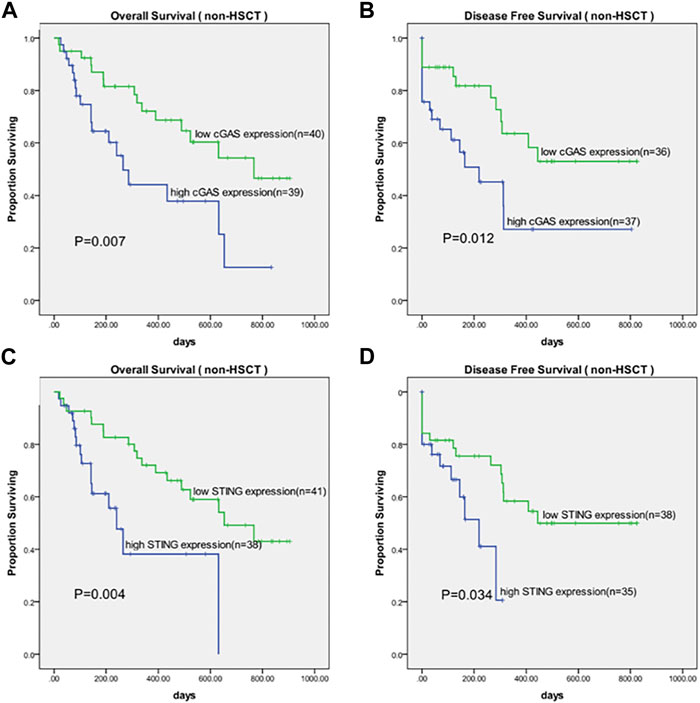
Figure 2. Kaplan-Meier survival curves of OS and DFS in patients grouped by median values of cGAS and STING expression. (A) Survival curves of OS in cGAS high and low groups. (B) Survival curves of DFS in cGAS high and low groups. (C) Survival curves of OS in STING high and low groups. (D) Survival curves of DFS in STING high and low groups. OS, overall survival; DFS, disease-free survival.
Univariate and multivariate analysis of factors affecting OS and DFS
The correlation between high cGAS and STING expression with inferior survival in AML patients indicated cGAS and STING expression levels might be of prognostic importance for AML. The results of univariate and multivariate analysis of factors affecting OS and DFS were shown in Table 2. The factors shown in patient characteristics were included in univariate analysis and variables with p < 0.1 were further analyzed in multivariate analysis. In multivariate Cox regression analysis, high cGAS expression was associated with inferior OS (HR = 2.348, 95% CI: 1.012–5.447; p = 0.047) and DFS (HR = 2.420, 95% CI: 1.071–5.469; p = 0.034). Nevertheless, it showed that there was no association between STING gene expression and OS or DFS. Besides, DNMT3a mutation, TET2 mutation were shown to be associated with worse OS and DFS while receiving allo-HSCT was associated with improved OS and DFS.
Positive correlation between cGAS and STING gene expression
As the above results showed, cGAS and STING expression was up-regulated in AML patients compared with normal controls. Although cGAS and SING are the entry of cGAS-STING pathway, previous study showed cGAS and STING expression could be regulated inconsistently in NSCLC [19]. We wondered whether cGAS and STING were up-regulated in a synchronized manner on the context of AML. Our result showed that in AML, cGAS expression was positively correlated with STING expression (R = 0.77, 95% CI: 0.6884–0.8301, p < 0.0001, Figure 3).
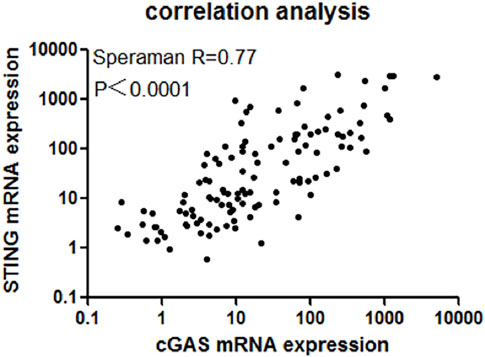
Figure 3. There existed a strong positive correlation between cGAS and STING expression (R = 0.77, 95% CI: 0.6884–0.8301, p < 0.0001).
Discussion
AML is a highly genetically heterogeneous malignant myeloproliferative disorder of bone marrow, accounting for ∼10% of all hematological diseases [1, 20]. While the next-generation sequencing technology has been tremendously developed, numerous recurrent point mutations, epigenetic changes as well as cytogenetic abnormality have been thoroughly recognized [21, 22]. Cytogenetics combined with mutations form the basis of the risk classification system, which facilitates the risk stratification for patients. However, up to 50% patients have been diagnosed as intermediate risk AML with a wide range of clinical outcomes. Thus, the identification of vital mechanisms affecting AML management and patient survival may boost the development of AML specific targeted therapies and meticulous risk stratification.
The results from current study indicated that high cGAS and STING expression correlated to inferior survival in AML. In multivariate analysis, only cGAS expression was found to be an independent factor affecting OS and DFS. It was intriguing that the expression level of STING showed impact on OS but not on DFS. Since our result showed cGAS and STING had a strong positive correlation, there might exist an overlapped effect of cGAS and STING expression on the clinical outcomes. Besides, recent work has demonstrated that cGAS and STING may act in an independent way from one another. Upon etoposide-induced DNA damage, STING could be activated independently from the catalytic function of cGAS [23]. The exquisite molecular mechanism defining the acting pattern of cGAS and STING in AML remained undetermined. In this regard, future studies are needed to investigate whether combination of cGAS and STING expression detection is necessary and cGAS gene expression alone can be an indicator for prognosis in AML.
Previous studies indicated the activation of cGAS-STING pathway contributed to cancer suppression by promoting host immuno-surveillance and inducing cellular senescence [24–27]. In line with these findings, cGAS-STING pathway was identified as a prognostic biomarker for improved clinical outcomes in hepatocellular carcinoma and non-small cell lung cancer (NSCLC) [19, 28]. However, other studies revealed that cGAS-STING pathway promoted tumor development and progression in Lewis lung carcinoma, brain and colorectal cancer [14, 15, 29]. One possible mechanism was that chronic stimulation of cGAS-STING pathway might lead to inflammation-driven carcinogenesis [30]. The topic that pro-inflammatory mediators are linked to AML cell growth has gain traction in recent years. Plenty of studies reported that chronic immune-stimulatory or autoimmune disease could be sick factors for developing AML [31, 32]. Here, our results suggested that cGAS-STING pathway also had a potential role in driving malignant programs and suppressing antitumor functions in AML. Whether hyper-activation of cGAS-STING pathway fueled the progression of AML via induction of inflammation requires further exploration. Also, future studies will be required to elucidate mechanism of hyper-activation cGAS-STING signaling in AML.
At present, NRAS/KRAS mutations are widely considered to be associated with poor prognosis in a variety of cancers, including colorectal cancer and serous ovarian cancer [33, 34]. NRAS mutations were identified in 10%–11% of AML, and KRAS mutations in an additional 5% [35, 36], however, how it affects cGAS-STING levels in AML has not been reported. Only one study in KRAS-mutant non-small-cell lung carcinoma lung cancer (NSCLC) may be relevant. In this study, the authors found that NRF2 promotes the transcription and expression of BRCA1 to repair DNA damage, leading to inactivation of the STING pathway [37]. In our study, the frequency of NRAS/KRAS mutation was higher in both cGAS and STING high group. TBK1 is the important downstream effector program of cGAS-STING signaling [7]. Previous work presented that TBK1 supported key, context specific tumorigenic activity in Ras-mutant/mesenchymal NSCLC [38]. The interaction between cGAS-STING pathway including its downstream signaling with NRAS/KRAS mutation on the context of AML would be an interesting area of research. Additionally, we found that elevated expression of cGAS was associated with higher risk stratification and a lower CR rate. Chromosomal instability (CIN) is a hallmark of cancer as well as a primary source of cytosolic dsDNA and it promotes the activation of cGAS-STING [39]. Intriguingly, some scholars found that cGAS could exert the function of maintaining CIN, which potentiated tumor evolution [40, 41]. In our study, AML patients had shown hyper-activation of the cGAS-STING pathway before treatment. The following chemotherapy further induced CIN, which might cooperate with cGAS-STING, leading to further perturbation of this pathway. Yet, the underlying mechanisms that define the effect of cGAS-STING pathway on treatment response in AML patients need further exploration.
In summary, our data revealed a prognosis role of cGAS-STING pathway for clinical outcomes and a positive correlation between cGAS and STING in AML. However, we recognize the limitations to our study, including the limited number of patients enrolled and the lack of relevant mechanism studied. In addition, the cGAS-STING downstream signaling programs including TBK1, IRF3, JAK2/STAT3, NF-κB were not studied in this study. Besides, it is important to note that the dichotomous roles of cGAS-STING in tumor immunity and development are cell and context - dependent [42]. Thus, in future study, a pinpoint cGAS-STING expression state should be better characterized in both immune cells and non-immune cells including tumor cells as well as stromal cell resident in bone marrow environment of AML patients. Despite various approaches for AML investigated, treatment resistance remains a leading cause of AML-related deaths [43]. Based on the results of this study, further studies of novel approaches targeting the cGAS-STING pathway in AML may provide potential for advancing AML therapeutic strategies.
Author contributions
Conceptualization, QLC and YH; Data curation, WFC, LZ, and YTH; Formal analysis, JWY, YH, and BX; Funding acquisition, FL, JWY, and BX; Methodology, QLC, JWZ, and WFC; Resources, QLC and BX; Software, FL, YTH, and BX; Supervision, JWY and BX; Validation, QLC and LZ; Writing the Original draft, JWY, QLC, and YH; Reviewing and editing the manuscript, JWY, QLC, and YH.
Data availability statement
The original contributions presented in the study are included in the article/supplementary material, further inquiries can be directed to the corresponding authors.
Ethics statement
The studies involving humans were approved by the Ethics Committee of The First Affiliate Hospital of Xiamen University (protocol code KY-2018-015 and 7 March 2018 of approval). The studies were conducted in accordance with the local legislation and institutional requirements. The participants provided their written informed consent to participate in this study.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was supported by the National Natural Science Foundation of China (81770126, 81770161, and 81800163), the Xiamen Municipal Bureau of Science and Technology (No. 3502Z20224011, 3502Z20227346, and 3502Z20227261) and the Natural Science Foundation of Xiamen City (20231301).
Publisher’s note
Please note that the review of this paper was conducted at the previous publisher, SAGE.
Footnotes
2http://tcga-data.nci.nih.gov/tcga/
3http://www.gtexportal.org/home/index.html
4http://www.ncbi.nlm.nih.gov/geo
References
1. Estey, E, and Döhner, H. Acute myeloid leukaemia. The Lancet (2006) 368:1894–907. doi:10.1016/s0140-6736(06)69780-8
2. Grimwade, D, Walker, H, Oliver, F, Wheatley, K, Harrison, C, Harrison, G, et al. The importance of diagnostic cytogenetics on outcome in AML: analysis of 1,612 patients entered into the MRC AML 10 trial. Blood (1998) 92:2322–33. doi:10.1182/blood.v92.7.2322.2322_2322_2333
3. Byrd, JC, Mrozek, K, Dodge, RK, Carroll, AJ, Edwards, CG, Arthur, DC, et al. Pretreatment cytogenetic abnormalities are predictive of induction success, cumulative incidence of relapse, and overall survival in adult patients with de novo acute myeloid leukemia: results from Cancer and Leukemia Group B (CALGB 8461). Blood (2002) 100:4325–36. doi:10.1182/blood-2002-03-0772
4. Ho, T-C, LaMere, M, Stevens, BM, Ashton, JM, Myers, JR, O'Dwyer, KM, et al. Evolution of acute myelogenous leukemia stem cell properties after treatment and progression. Blood (2016) 128:1671–8. doi:10.1182/blood-2016-02-695312
5. Ishikawa, F, Yoshida, S, Saito, Y, Hijikata, A, Kitamura, H, Tanaka, S, et al. Chemotherapy-resistant human AML stem cells home to and engraft within the bone-marrow endosteal region. Nat Biotechnol (2007) 25:1315–21. doi:10.1038/nbt1350
6. Estey, EH. Acute myeloid leukemia: 2019 update on risk-stratification and management. Am J Hematol (2018) 93:1267–91. doi:10.1002/ajh.25214
7. Zhang, X, Bai, XC, and Chen, ZJ. Structures and mechanisms in the cGAS-STING innate immunity pathway. Immunity (2020) 53:43–53. doi:10.1016/j.immuni.2020.05.013
8. Hopfner, KP, and Hornung, V. Molecular mechanisms and cellular functions of cGAS-STING signalling. Nat Rev Mol Cell Biol (2020) 21:501–21. doi:10.1038/s41580-020-0244-x
9. Kwon, J, and Bakhoum, SF. The cytosolic DNA-sensing cGAS-STING pathway in cancer. Cancer Discov (2020) 10:26–39. doi:10.1158/2159-8290.cd-19-0761
10. Curran, E, Chen, X, Corrales, L, Kline, DE, Dubensky, TW, Duttagupta, P, et al. STING pathway activation stimulates potent immunity against acute myeloid leukemia. Cell Rep (2016) 15:2357–66. doi:10.1016/j.celrep.2016.05.023
11. Kogan, AA, Topper, MJ, Dellomo, AJ, Stojanovic, L, McLaughlin, LJ, Creed, TM, et al. Activating STING1-dependent immune signaling in TP53 mutant and wild-type acute myeloid leukemia. Proc Natl Acad Sci U S A (2022) 119:e2123227119. doi:10.1073/pnas.2123227119
12. Song, C, Liu, D, Liu, S, Li, D, Horecny, I, Zhang, X, et al. SHR1032, a novel STING agonist, stimulates anti-tumor immunity and directly induces AML apoptosis. Sci Rep (2022) 12:8579. doi:10.1038/s41598-022-12449-1
13. Song, X, Peng, Y, Wang, X, Chen, Q, Lan, X, and Shi, F. The stimulator of interferon genes (STING) agonists for treating acute myeloid leukemia (AML): current knowledge and future outlook. Clin Transl Oncol (2022) 25:1545–53. doi:10.1007/s12094-022-03065-6
14. Chen, Q, Boire, A, Jin, X, Valiente, M, Er, EE, Lopez-Soto, A, et al. Carcinoma-astrocyte gap junctions promote brain metastasis by cGAMP transfer. Nature (2016) 533:493–8. doi:10.1038/nature18268
15. Lemos, H, Mohamed, E, Huang, L, Ou, R, Pacholczyk, G, Arbab, AS, et al. STING promotes the growth of tumors characterized by low antigenicity via IDO activation. Cancer Res (2016) 76:2076–81. doi:10.1158/0008-5472.can-15-1456
16. Ahn, J, Xia, T, Konno, H, Konno, K, Ruiz, P, and Barber, GN. Inflammation-driven carcinogenesis is mediated through STING. Nat Commun (2014) 5:5166. doi:10.1038/ncomms6166
17. Tang, Z, Li, C, Kang, B, Gao, G, Li, C, and Zhang, Z. GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res (2017) 45:W98–w102. doi:10.1093/nar/gkx247
18. Barrett, T, Wilhite, SE, Ledoux, P, Evangelista, C, Kim, IF, Tomashevsky, M, et al. NCBI GEO: archive for functional genomics data sets--update. Nucleic Acids Res (2012) 41:D991–5. doi:10.1093/nar/gks1193
19. Raaby Gammelgaard, K, Sandfeld-Paulsen, B, Godsk, SH, Demuth, C, Meldgaard, P, Sorensen, BS, et al. cGAS-STING pathway expression as a prognostic tool in NSCLC. Transl Lung Cancer Res (2021) 10:340–54. doi:10.21037/tlcr-20-524
20. Döhner, H, Estey, EH, Amadori, S, Appelbaum, FR, Büchner, T, Burnett, AK, et al. Diagnosis and management of acute myeloid leukemia in adults: recommendations from an international expert panel, on behalf of the European LeukemiaNet. Blood (2010) 115:453–74. doi:10.1182/blood-2009-07-235358
21. Ley, TJ, Mardis, ER, Ding, L, Fulton, B, McLellan, MD, Chen, K, et al. DNA sequencing of a cytogenetically normal acute myeloid leukaemia genome. Nature (2008) 456:66–72. doi:10.1038/nature07485
22. Care, MA, Barrans, S, Worrillow, L, Jack, A, Westhead, DR, and Tooze, RM. A microarray platform-independent classification tool for cell of origin class allows comparative analysis of gene expression in diffuse large B-cell lymphoma. PLoS One (2013) 8:e55895. doi:10.1371/journal.pone.0055895
23. Dunphy, G, Flannery, SM, Almine, JF, Connolly, DJ, Paulus, C, Jønsson, KL, et al. Non-canonical activation of the DNA sensing adaptor STING by ATM and IFI16 mediates NF-κB signaling after nuclear DNA damage. Mol Cell (2018) 71:745–60.e5. doi:10.1016/j.molcel.2018.07.034
24. Yang, H, Wang, H, Ren, J, Chen, Q, and Chen, ZJ. cGAS is essential for cellular senescence. Proc Natl Acad Sci U S A (2017) 114:E4612–e20. doi:10.1073/pnas.1705499114
25. Glück, S, Guey, B, Gulen, MF, Wolter, K, Kang, TW, Schmacke, NA, et al. Innate immune sensing of cytosolic chromatin fragments through cGAS promotes senescence. Nat Cell Biol (2017) 19:1061–70. doi:10.1038/ncb3586
26. Diamond, MS, Kinder, M, Matsushita, H, Mashayekhi, M, Dunn, GP, Archambault, JM, et al. Type I interferon is selectively required by dendritic cells for immune rejection of tumors. J Exp Med (2011) 208:1989–2003. doi:10.1084/jem.20101158
27. Ahn, J, Konno, H, and Barber, GN. Diverse roles of STING-dependent signaling on the development of cancer. Oncogene (2015) 34:5302–8. doi:10.1038/onc.2014.457
28. Qi, Z, Yan, F, Chen, D, Xing, W, Li, Q, Zeng, W, et al. Identification of prognostic biomarkers and correlations with immune infiltrates among cGAS-STING in hepatocellular carcinoma. Biosci Rep (2020) 40:BSR20202603. doi:10.1042/BSR20202603
29. An, X, Zhu, Y, Zheng, T, Wang, G, Zhang, M, Li, J, et al. An analysis of the expression and association with immune cell infiltration of the cGAS/STING pathway in pan-cancer. Mol Ther - Nucleic Acids (2019) 14:80–9. doi:10.1016/j.omtn.2018.11.003
30. Khoo, LT, and Chen, LY. Role of the cGAS-STING pathway in cancer development and oncotherapeutic approaches. EMBO Rep (2018) 19:e46935. doi:10.15252/embr.201846935
31. Ramadan, SM, Fouad, TM, Summa, V, Hasan, SK, and Lo-Coco, F. Acute myeloid leukemia developing in patients with autoimmune diseases. Haematologica (2012) 97:805–17. doi:10.3324/haematol.2011.056283
32. Carey, A, Edwards, DK, Eide, CA, Newell, L, Traer, E, Medeiros, BC, et al. Identification of interleukin-1 by functional screening as a key mediator of cellular expansion and disease progression in acute myeloid leukemia. Cell Rep (2017) 18:3204–18. doi:10.1016/j.celrep.2017.03.018
33. De Roock, W, Claes, B, Bernasconi, D, De Schutter, J, Biesmans, B, Fountzilas, G, et al. Effects of KRAS, BRAF, NRAS, and PIK3CA mutations on the efficacy of cetuximab plus chemotherapy in chemotherapy-refractory metastatic colorectal cancer: a retrospective consortium analysis. Lancet Oncol (2010) 11:753–62. doi:10.1016/s1470-2045(10)70130-3
34. Emmanuel, C, Chiew, YE, George, J, Etemadmoghadam, D, Anglesio, MS, Sharma, R, et al. Genomic classification of serous ovarian cancer with adjacent borderline differentiates RAS pathway and TP53-mutant tumors and identifies NRAS as an oncogenic driver. Clin Cancer Res (2014) 20:6618–30. doi:10.1158/1078-0432.ccr-14-1292
35. Bacher, U, Haferlach, T, Schoch, C, Kern, W, and Schnittger, S. Implications of NRAS mutations in AML: a study of 2502 patients. Blood (2006) 107:3847–53. doi:10.1182/blood-2005-08-3522
36. Bowen, DT, Frew, ME, Hills, R, Gale, RE, Wheatley, K, Groves, MJ, et al. RAS mutation in acute myeloid leukemia is associated with distinct cytogenetic subgroups but does not influence outcome in patients younger than 60 years. Blood (2005) 106:2113–9. doi:10.1182/blood-2005-03-0867
37. Xiaodan, S, Peiyan, Z, Hui, L, Yan, L, and Ying, C. NRF2 participates in the suppressive tumor immune microenvironment of KRAS/KEAP1 co-mutant non-small cell lung cancer by inhibiting the STING pathway. Genes Dis (2023) 10:1727–30. doi:10.1016/j.gendis.2022.10.009
38. Cooper, JM, Ou, YH, McMillan, EA, Vaden, RM, Zaman, A, Bodemann, BO, et al. TBK1 provides context-selective support of the activated AKT/mTOR pathway in lung cancer. Cancer Res (2017) 77:5077–94. doi:10.1158/0008-5472.can-17-0829
39. Wan, D, Jiang, W, and Hao, J. Research advances in how the cGAS-STING pathway controls the cellular inflammatory response. Front Immunol (2020) 11:615. doi:10.3389/fimmu.2020.00615
40. Bakhoum, SF, and Cantley, LC. The multifaceted role of chromosomal instability in cancer and its microenvironment. Cell (2018) 174:1347–60. doi:10.1016/j.cell.2018.08.027
41. Liu, H, Zhang, H, Wu, X, Ma, D, Wu, J, Wang, L, et al. Nuclear cGAS suppresses DNA repair and promotes tumorigenesis. Nature (2018) 563:131–6. doi:10.1038/s41586-018-0629-6
42. Ng, KW, Marshall, EA, Bell, JC, and Lam, WL. cGAS-STING and cancer: dichotomous roles in tumor immunity and development. Trends Immunol (2018) 39:44–54. doi:10.1016/j.it.2017.07.013
Keywords: cGAS, STING, expression, prognosis, acute myeloid leukemia
Citation: Chen Q, Hong Y, Chen W, Lin F, Zeng J, Huang Y, Zhang L, Yao J and Xu B (2024) Prognostic implications of cGAS and STING gene expression in acute myeloid leukemia. Exp. Biol. Med. 249:10108. doi: 10.3389/ebm.2024.10108
Received: 09 July 2023; Accepted: 25 September 2023;
Published: 29 February 2024.
Copyright © 2024 Chen, Hong, Chen, Lin, Zeng, Huang, Zhang, Yao and Xu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Bing Xu, eHViaW5nQHhtdS5lZHUuY24=; Jingwei Yao, MzgxMDYwOTg1QHFxLmNvbQ==; Li Zhang, OTU3ODM3MjQyQHFxLmNvbQ==
†These authors have contributed equally to this work
 Qiuling Chen1,2†
Qiuling Chen1,2† Bing Xu
Bing Xu